Truncated variant of CRISPR nuclease SpCas9 of streptococcus pyogenes and application of truncated variant
A nuclease, nucleic acid technology, applied in the field of protein engineering, can solve the problems of small loading, complex preparation, low titer and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1, constructing the plasmid of CRISPR-Cas9 nuclease (TSpCas9).
[0055] 1. Mutant Design.
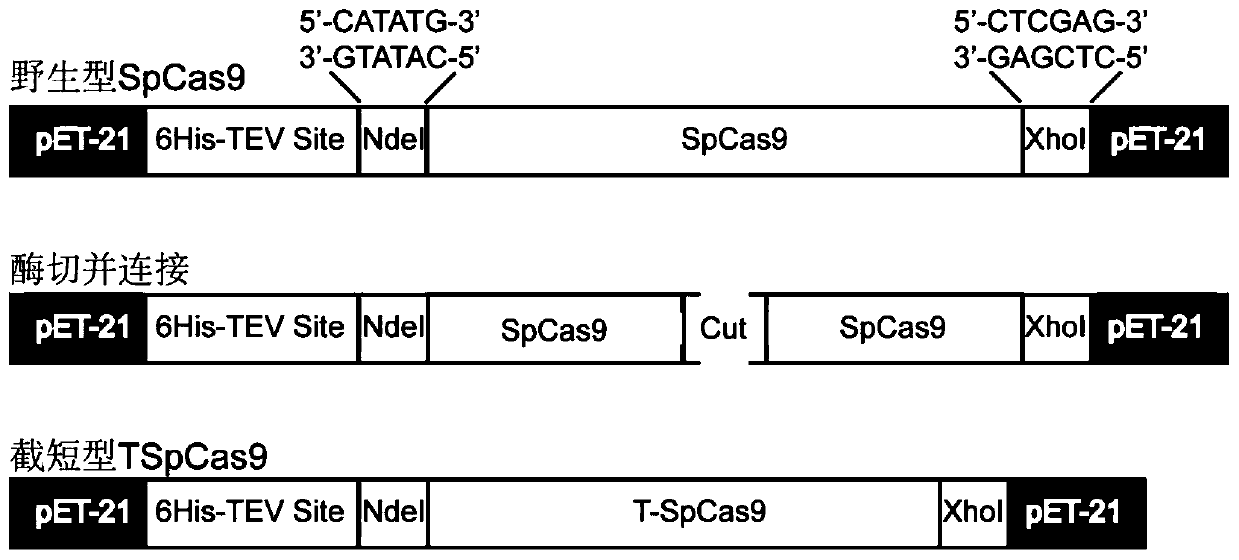
[0056] Using the pet21-6His-TEV-SpCas9 plasmid, namely SEQ ID NO.5, as a template, truncated 180aa (5822bp) to 299aa (6181bp), which is equivalent to the base sequence of SEQ ID NO.8 and the amino acid sequence of SEQ ID NO.9, The remaining part was recombined into a truncated SpCas9, which was called TSpCas9. Its transformation design idea is as follows: figure 1 As shown, the detailed steps are briefly described as follows:
[0057] First, use primers F-F and F-R, S-F and S-R to amplify 5267~5821bp (equivalent to 1-538bp of SEQ ID NO.10) and 6182~9391bp (equivalent to 898-4104bp of SEQ ID NO.12), and passed AxyPrep TM DNA Gel Extraction Kit (purchased from Axygen) was purified and recovered, and they were called amplified fragments F and S;
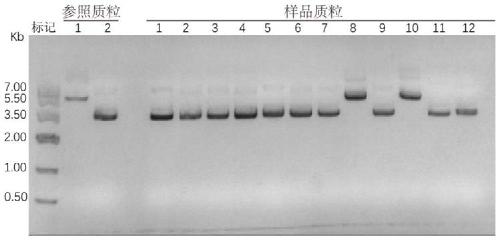
[0058] Secondly, use restriction endonucleases NdeI and XhoI (purchased from NEB) to digest pet21-6His-TEV-SpCas9 and F amp...
Embodiment 2
[0096] Example 2, preparing CRISPR-Cas9 (TSpCas9) nuclease.
[0097] 2. Protein expression and purification.
[0098] 2.1 Protein expression
[0099] (1) Turn on the ultra-clean bench, wipe the table top and various utensils with a cotton ball containing 75% alcohol, turn on the ultraviolet light for 20 minutes, and start the fan for standby;
[0100] (2) Pipette 10 μl of Rosetta (DE3) (purchased from TIANGEN) bacterial solution expressing Pet21-6His-TEV-TSpCas9 into 6 ml of LB liquid medium containing double antibodies (Amp and Cm), 37°C, 200r / min shaking culture overnight;
[0101] (3) Transfer the overnight cultured bacterial solution to 500ml LB (purchased from Sanko) liquid medium containing double antibodies according to the volume ratio of 1:100, and cultivate at 37°C with shaking at 200r / min. During the cultivation process, detect the OD value of the bacterial solution at any time;
[0102] (4) When the OD value of the bacterial solution is close to 0.4-0.8, add t...
Embodiment 3
[0112] Example 3, testing CRISPR-Cas9 (TSpCas9) nuclease cleavage activity.
[0113] 3. Detection of mutant activity.
[0114] The substrate DNA (SEQ ID NO.20) used was mainly obtained by conventional PCR amplification using primers QG-F: TAGTCCTGTCGGGTTTCG (SEQ ID NO.17) and QG-R: TTCCATTCGCCATTCAGG (SEQ ID NO.18). The reaction system and amplification conditions are as follows:
[0115] The amplification system is as follows:
[0116]
[0117] PCR reaction conditions:
[0118]
[0119] (3) Procurement of rubber tapping recovery kits
[0120] The rubber tapping recovery kit AxyPrep used TM The DNA Gel Extraction Kit was ordered from Axygen Company, and the rubber tapping recovery operation was carried out according to its instructions, and a relatively pure substrate DNA (SEQ ID NO.20) could be obtained.
[0121] Cas9 and sgRNA are mixed in equimolar amounts, and the substrate DNA can be adjusted to 0.2-1 times the molar mass of Cas9 according to experimental need...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com