Biomolecules mass spectrometry detection method
A technology for mass spectrometry detection and biomolecules, applied in the field of mass spectrometry detection of biomolecules, can solve the problems of limited fragmentation efficiency, limited fragmentation information, lack of fragmentation information, etc., to achieve improved sensitivity and accuracy, high secondary fragmentation efficiency, rich The effect of fragmented information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
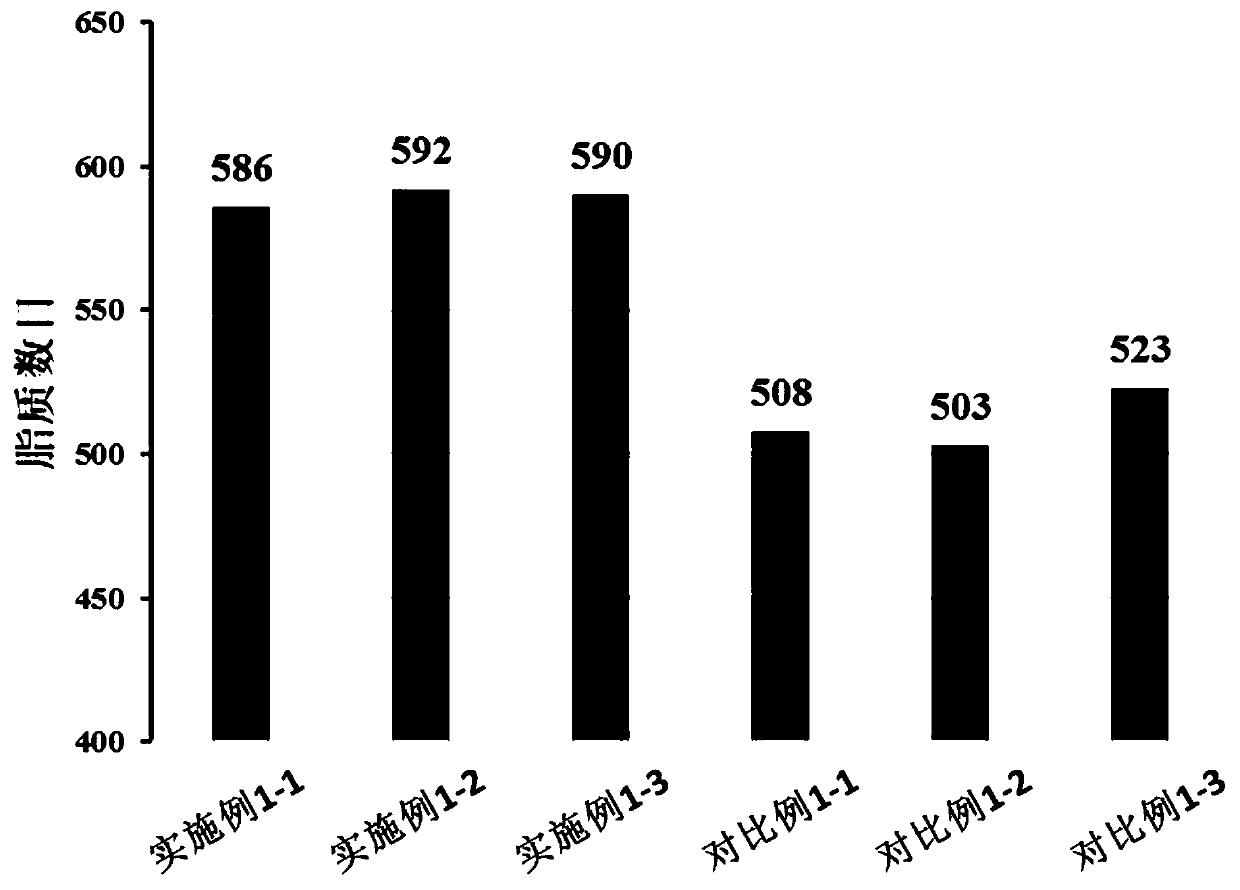
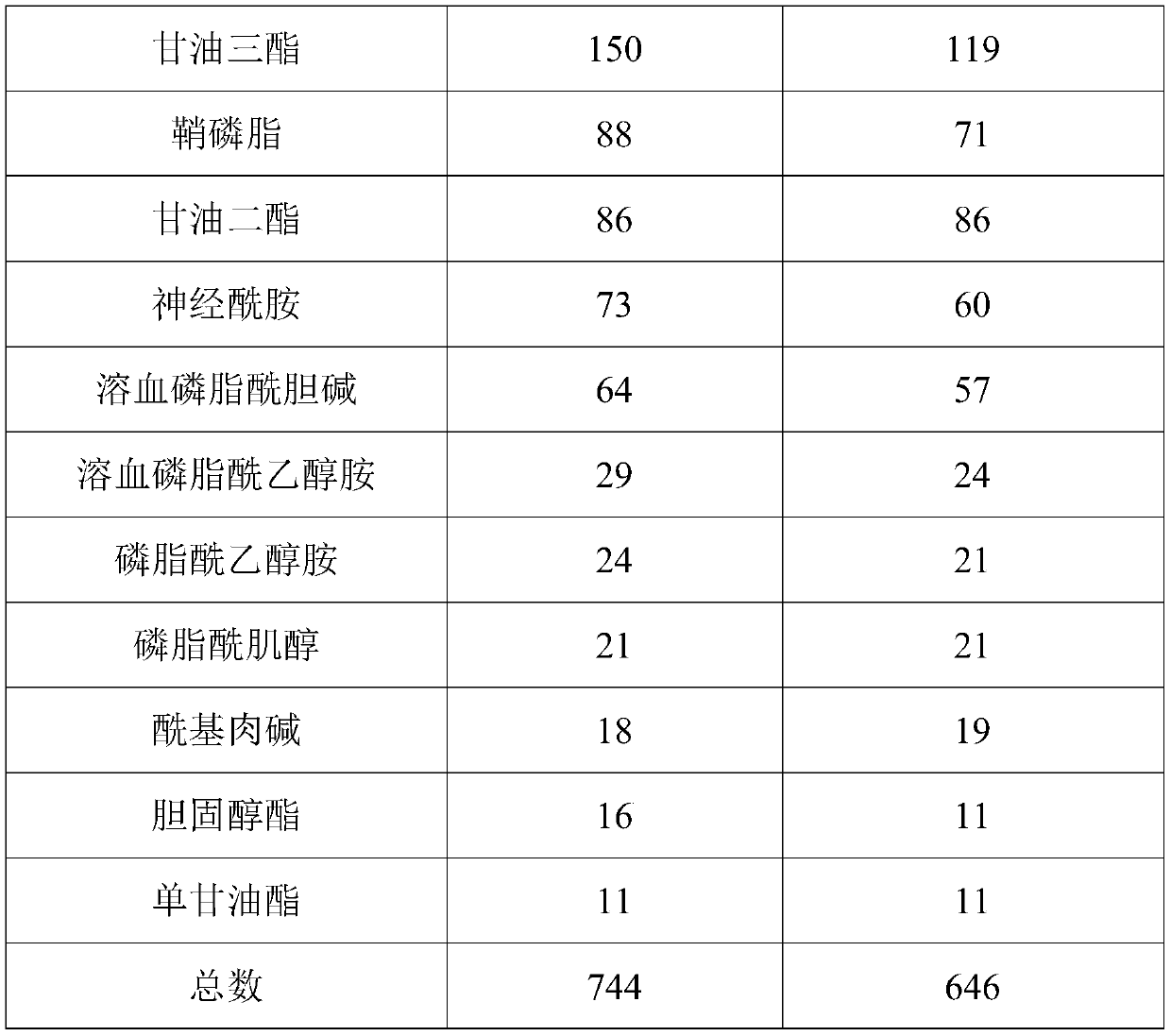
[0060] This embodiment provides a mass spectrometry detection method for lipidomics analysis, which specifically includes the following steps:
[0061] (1) Sample preparation: Take 100uL of serum in a 1.5mL centrifuge tube, add 300uL of -20°C pre-cooled isopropanol, vortex for 1min; place the mixture at room temperature for 10min, and then place it in a -20°C refrigerator overnight , to enhance protein precipitation; centrifuge at 4°C for 20 min, take the supernatant, and freeze-dry. The lyophilized sample was reconstituted with 100 uL of reconstitution solution (volume ratio of isopropanol, methanol, and water: 50:35:15) to obtain the serum lipidomics analysis sample to be tested.
[0062] (2) Mass spectrometry detection: use a high-performance liquid chromatography-quadrupole-electrostatic field orbitrap high-resolution mass spectrometry system to perform mass spectrometry detection on the serum lipidomics analysis sample obtained in step (1), and obtain mass spectrometry da...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| The inside diameter of | aaaaa | aaaaa |
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com