Three internal reference genes of acipenser dabryanus, primer development and stability evaluation technique
An internal reference gene, Dabry's sturgeon technology, applied in the field of molecular biology, can solve problems such as the lack of internal reference genes in Dabry's sturgeon, and achieve the effects of improving reliability and repeatability, improving detection efficiency, and improving credibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
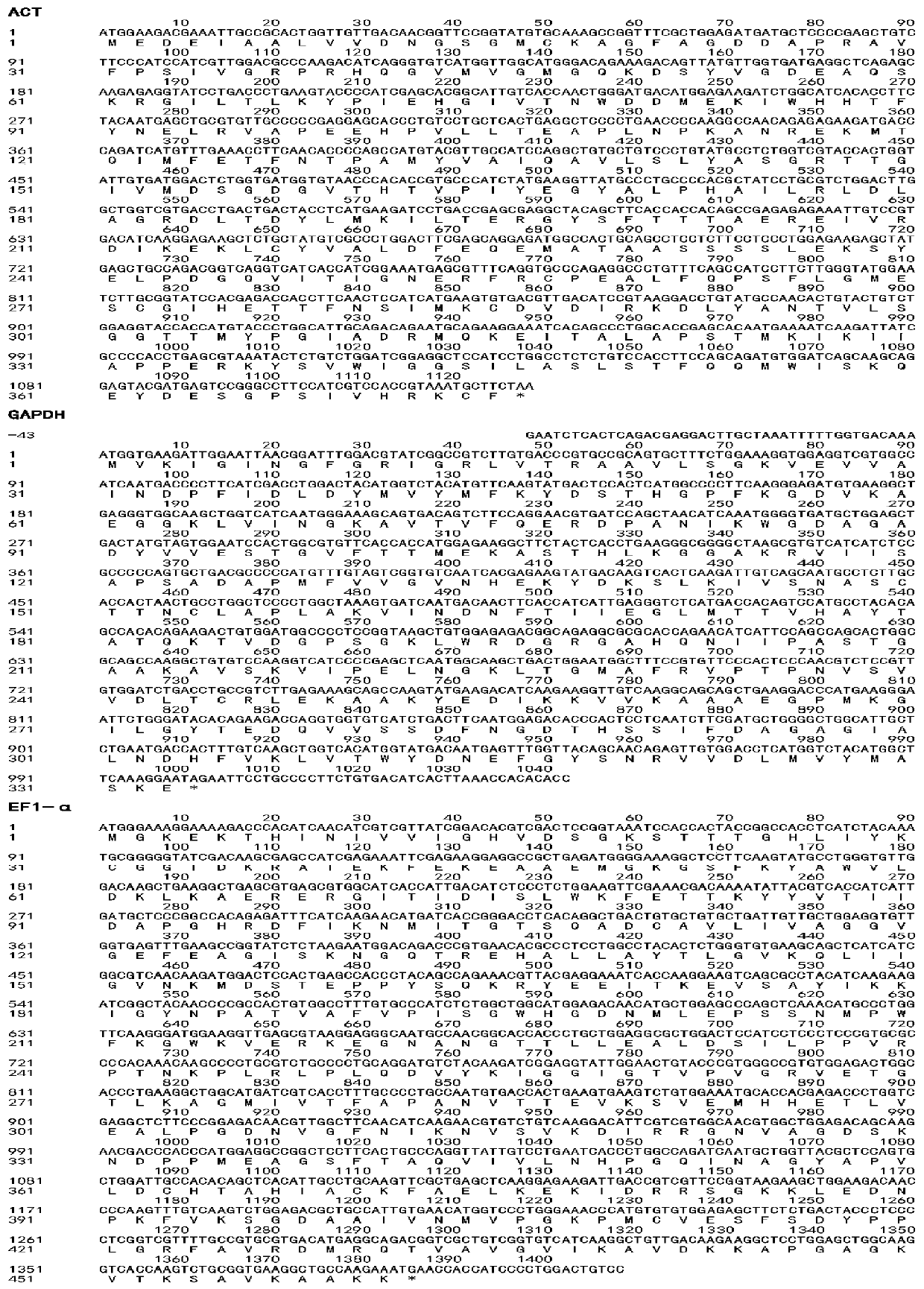
[0054] Example 1 Cloning of ACT, GAPDH and EF1-α genes of Dabry's sturgeon
[0055] 1) Design of primers for ACT, GAPDH and EF1-α genes
[0056] The ACT, GAPDH and EF1-α genes were cloned by homologous cloning method, searched the NCBI database and constructed the local Blast database, searched for the homologous sequences of the corresponding genes, and designed RT-PCR primers in the conserved regions of the homologous sequences. The partial sequence of the gene and the ACT gene of Acipenser dabryi were used to design primers for the ACT gene of Acipenser dabryi, and the GAPDH of Acipenser dabryi. Two sequences GETX01028036.1 and GEUL01069530.1 from Acipenser dabryii, Siberian sturgeon and local database were used to design primers for the EF1-α gene of Acipenser dabryi;
[0057] 2) Extraction of total RNA from the liver of Acipenser dabryi
[0058] Take about 100mg of liver tissue and place it in a 1.5ml EP tube pre-added with 100μl Buffer Rlysis-AG lysate. After fully gri...
Embodiment 2
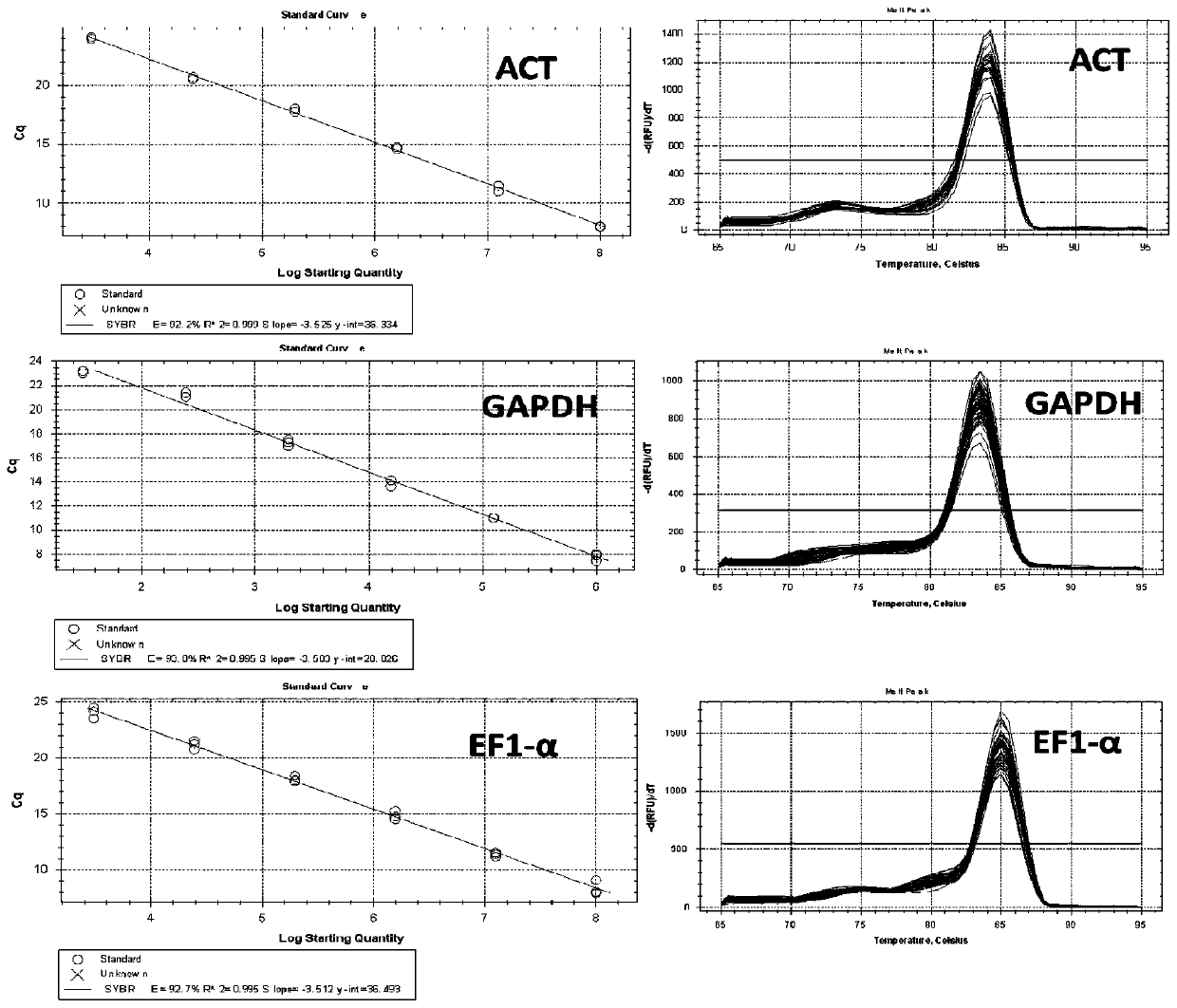
[0065] Example 2 Real-time fluorescent quantitative PCR primer design and primer quality detection
[0066] 1) Based on the nucleotide sequences of ACT, GAPDH and EF1-α obtained in Example 1, the fluorescent quantitative specific primers were designed according to the principles of real-time fluorescent quantitative PCR primer design, and the lengths of the amplified fragments were 203bp, 115bp and 171bp respectively , the fluorescent quantitative specific primers are the real-time fluorescent quantitative PCR primers, respectively
[0067] qACT-F CTGTTTCAGCCATCCTTCTTG
[0068] qACT-R TTGATTTTCATTGTGCTCGGT
[0069] qGAPDH-F TCAAATGGGGTGATGCTGGAG
[0070] qGAPDH-R GCGGAGATGATGACACGCTTAG
[0071] qEF1-α-F GAAAGGAAAAGACCCACAT
[0072] qEF1-α-R CCAGGCATACTTGAAGGAGC
[0073] 2) Extract the total RNA of Sturgeon dabryi's brain, and synthesize the first strand of cDNA according to the method of the PrimeScriptTM 1st Strand cDNA Synthesis Kit kit, that is, reverse transcribe the ...
Embodiment 3A
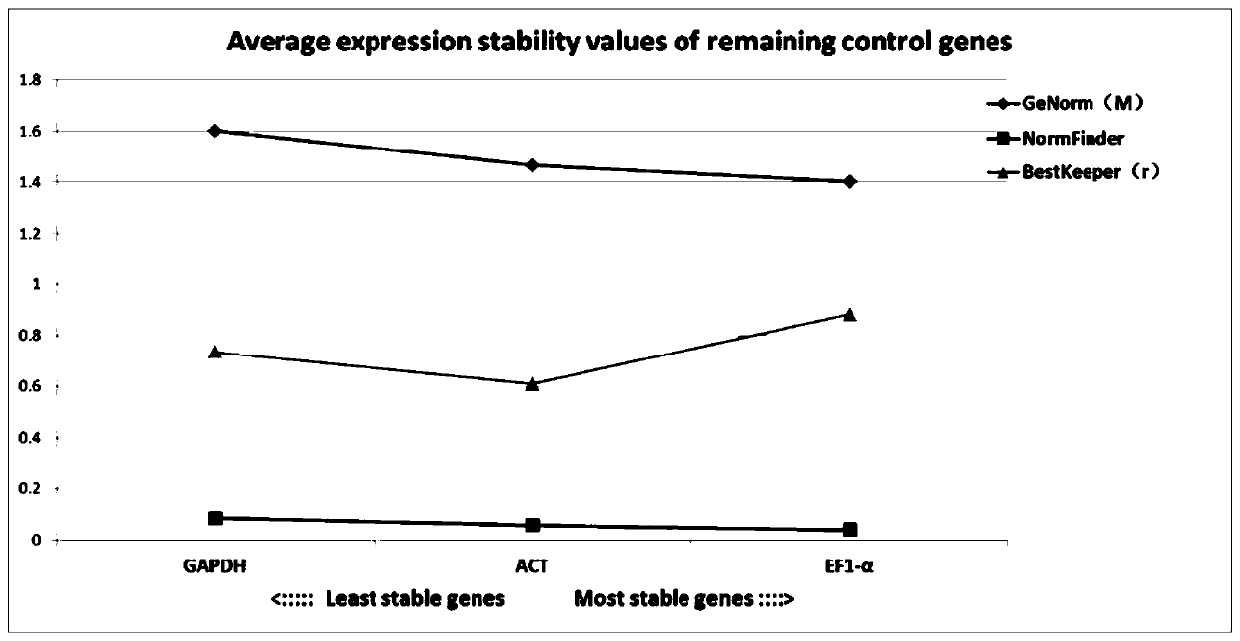
[0076] Tissue expression stability analysis of embodiment 3 ACT, GAPDH and EF1-α
[0077] 1) Tissue sampling
[0078] A 350±10g sample of three-tailed Dabry's sturgeon was collected at the Yibin Artificial Breeding Base of the Fisheries Research Institute of the Sichuan Academy of Agricultural Sciences. The Dabry's sturgeon was quickly dissected in a biological safety cabinet, and the brain, liver, kidney, pancreas, spleen, esophagus, stomach, and pylorus were removed. The caecum, duodenum, valve intestine, rectum, muscle, gills, and skin were quickly washed with ice-bathed saline to remove blood stains, and the processed liver was packed in a sample tube, which was quickly frozen in liquid nitrogen and stored at -80°C. ℃ refrigerator;
[0079] 2) RNA extraction
[0080] Take about 100mg of preserved tissue and place it in a 1.5ml EP tube pre-added with 100μl Buffer Rlysis-AG lysate. After fully grinding with an electric grinder, add 350μl of Buffer Rlysis-AG, and then refer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com