The internal reference gene of coral dish and its screening method and application
A technology of internal reference genes and screening methods, which is applied in the field of plant genetic engineering, can solve the problems of internal reference gene screening and stability verification without coral dish, and achieve the effects of improving scientific research work efficiency, improving calibration capabilities, and reducing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
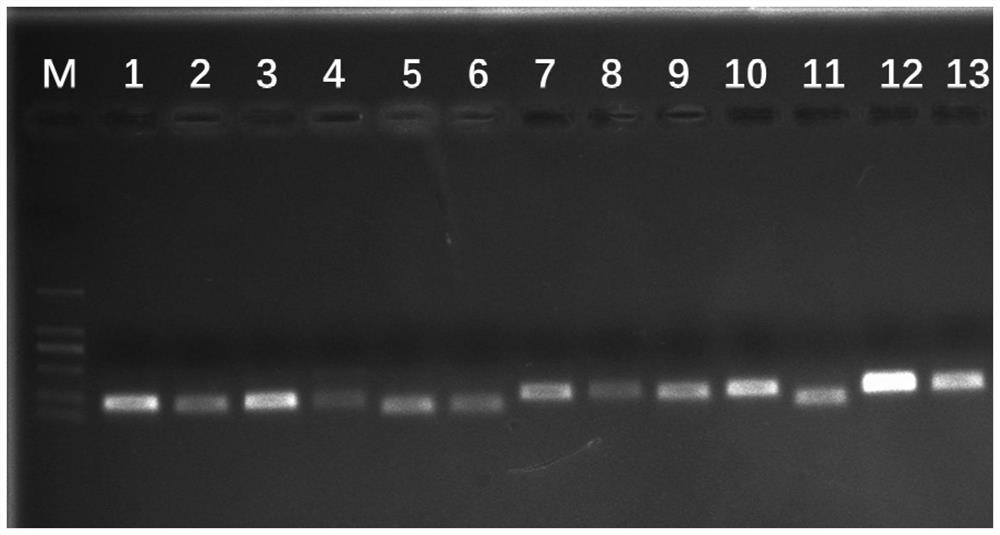
[0056] This example is an internal reference gene of the coral dish of the present invention. The following 13 internal reference genes of Corydalis are provided: PP2A (protein phosphatase 2), UBQ10 (ubiquitin 10), ACT (actin), EF1-α (elongation factor), GAPDH (glyceraldehyde-3-phosphate hydrogenase), α-TUB (α-tubulin), β-TUB (β-tubulin), PTBP1 (polypyrimidine sequence binding protein), EXP1 (expansin 1), EXP2 (expansin 2), TIP41-like (TIP41-like protein), SAND family (sand family protein), CYP2 (cyclophilin); the nucleotide sequence of the PP2A is SEQ ID NO.1; the nucleotide sequence of the UBQ10 is SEQ ID NO.2; the nucleotide sequence of the ACT is SEQ ID NO.3; the nucleotide sequence of the EF1-α is SEQ ID NO.4; the nucleotide sequence of the GAPDH is SEQ ID NO.5; The nucleotide sequence of the α-TUB is SEQ ID NO.6; the nucleotide sequence of the β-TUB is SEQ ID NO.7; the nucleotide sequence of the PTBP1 is SEQ ID NO.8; the nucleotide sequence of the β-TUB is SEQ ID NO.8; ...
Embodiment 2
[0058] In this example, fluorescent quantitative PCR primers for amplifying the internal reference gene in Example 1 are provided,
[0059] The size of the amplified fragment of the primers is 100-250bp.
[0060] The primer pair sequence used to amplify PP2A is SEQ ID NO.15 / SEQ ID NO.16; the primer pair sequence used to amplify UBQ10 is SEQ ID NO.17 / SEQ ID NO.18; the primer pair sequence used to amplify ACT The primer pair sequence is SEQ ID NO.19 / SEQID NO.20; the primer pair sequence for amplifying EF1-α is SEQ ID NO.21 / SEQ ID NO.22; the primer pair sequence for amplifying GAPDH is SEQ ID NO.21 / SEQ ID NO.22; ID NO.23 / SEQ ID NO.24; The primer pair sequence for amplifying α-TUB is SEQ ID NO.25 / SEQ ID NO.26; The primer pair sequence for amplifying β-TUB is SEQ ID NO. 27 / SEQ ID NO.28; the primer pair sequence used to amplify PTBP1 is SEQ ID NO.29 / SEQ ID NO.30; the primer pair sequence used to amplify EXP1 is SEQ ID NO.31 / SEQ ID NO. 32; the primer pair sequence used to amplify E...
Embodiment 3
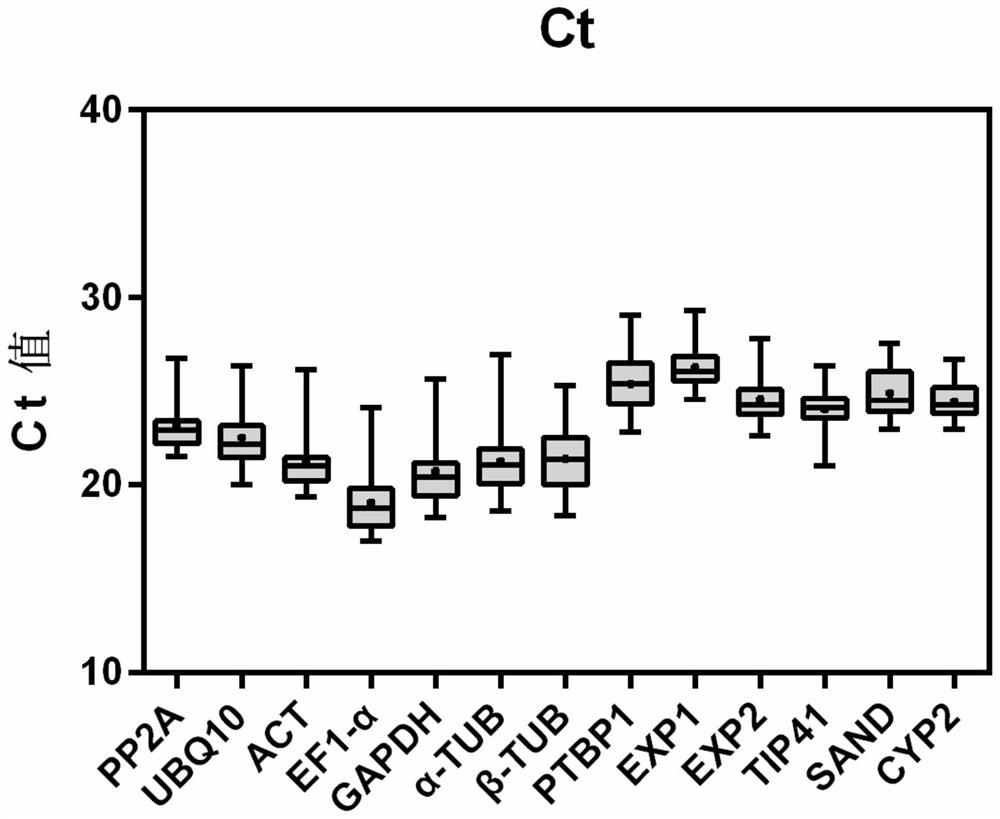
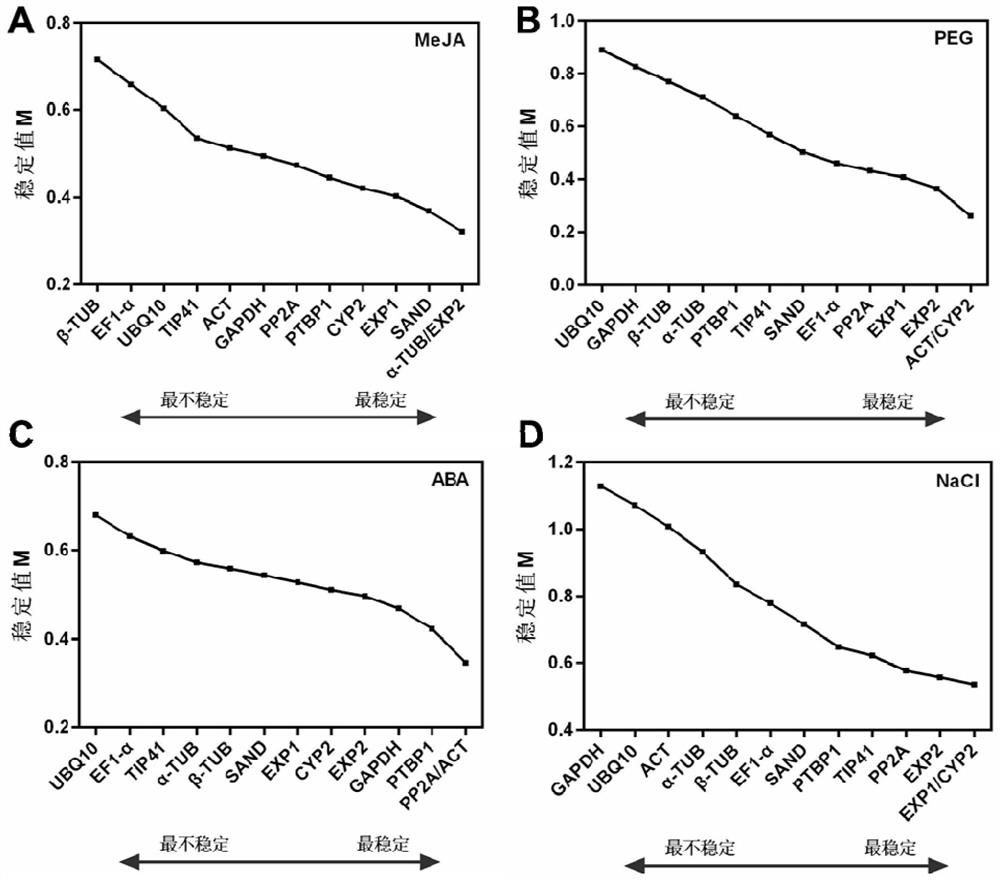
[0062] The present embodiment provides a real-time fluorescent quantitative PCR method for screening the internal reference genes of the above-mentioned Anchovy, which comprises the following steps: (1) selecting root tissues of Anchophylla that are respectively treated with salt stress, drought stress, abscisic acid, and methyl jasmonate sample;
[0063] (2) Use the sequencing data of the transcriptome of the coral dish to screen out the candidate internal reference genes of the coral dish;
[0064] (3) Use the selected candidate internal reference genes as templates (i.e. PP2A, UBQ10, ACT, EF1-α, GAPDH, α-TUB, β-TUB, PTBP1, EXP1, EXP2, TIP41-like, SAND family, CYP2; specific For the sequence, please refer to the sequence table (SEQ ID NO.1-SEQ ID NO.13), design and obtain the primers for real-time fluorescent quantitative PCR of each internal reference gene (see Example 2 for details);
[0065] (4) Perform real-time fluorescent quantitative PCR, perform statistical analysis...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com