Formate dehydrogenase mutant with improved substrate affinity and coenzyme affinity
A formate dehydrogenase and mutant technology, applied in the field of bioengineering, can solve the problems of high concentration of formate substrate and coenzyme, increased cost of coenzyme regeneration and reaction liquid treatment, unsuitable for practical application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1 Site-directed mutation of BstFDH
[0071] Through Uniprot, NCBI BLAST and spatial structure modeling, the three-dimensional spatial structure of BstFDH of the amino acid sequence shown in SEQ ID No. 2 in the sequence table was obtained, and the amino acid residues around the binding site of the coenzyme and the substrate were mutated. Use the recombinant plasmid of parent BstFDH as PCR template for site-directed saturation mutation. The PCR system is: 2×PrimeStar 10μl, upstream primer and downstream primer (10ng / μl) each 1μl, template plasmid (50ng / μl) 1μl, DMSO 1μl and ddH 2 O 6μl. The PCR amplification program is: pre-denaturation at 98°C for 5 minutes and then 30 cycles as follows: denaturation at 98°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 7 minutes; finally, extension at 72°C for 10 minutes. The PCR amplified product was added to DpnI for digestion for 2 hours, and the digested product was transformed into E.coli BL21(DE3...
Embodiment 2B
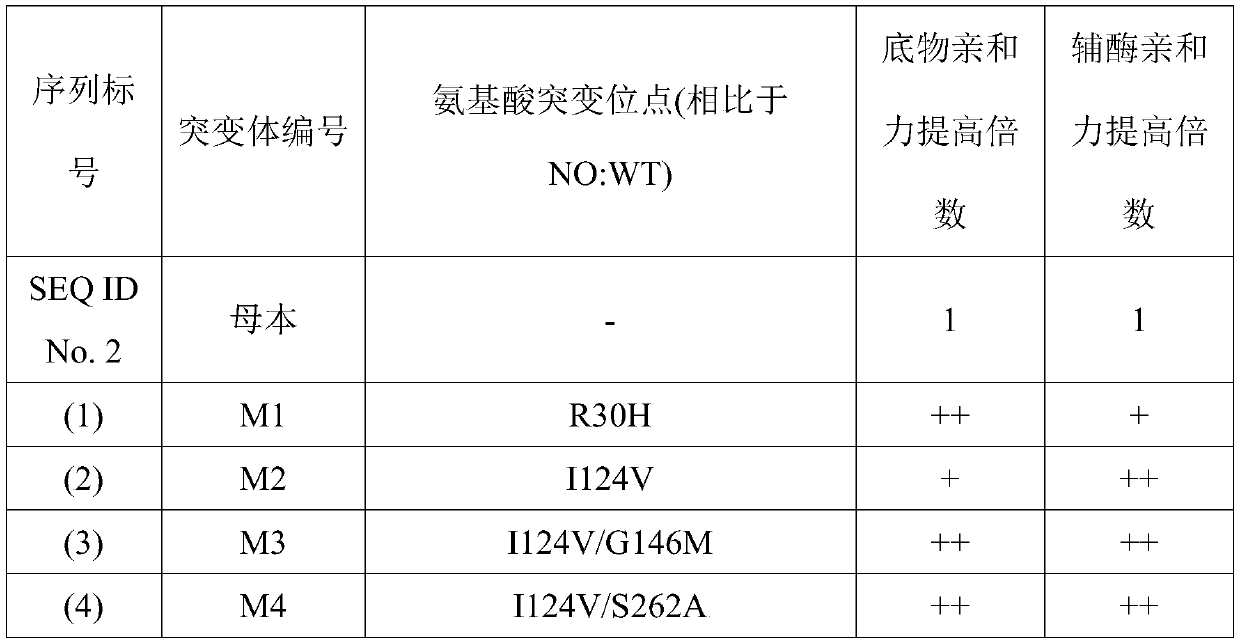
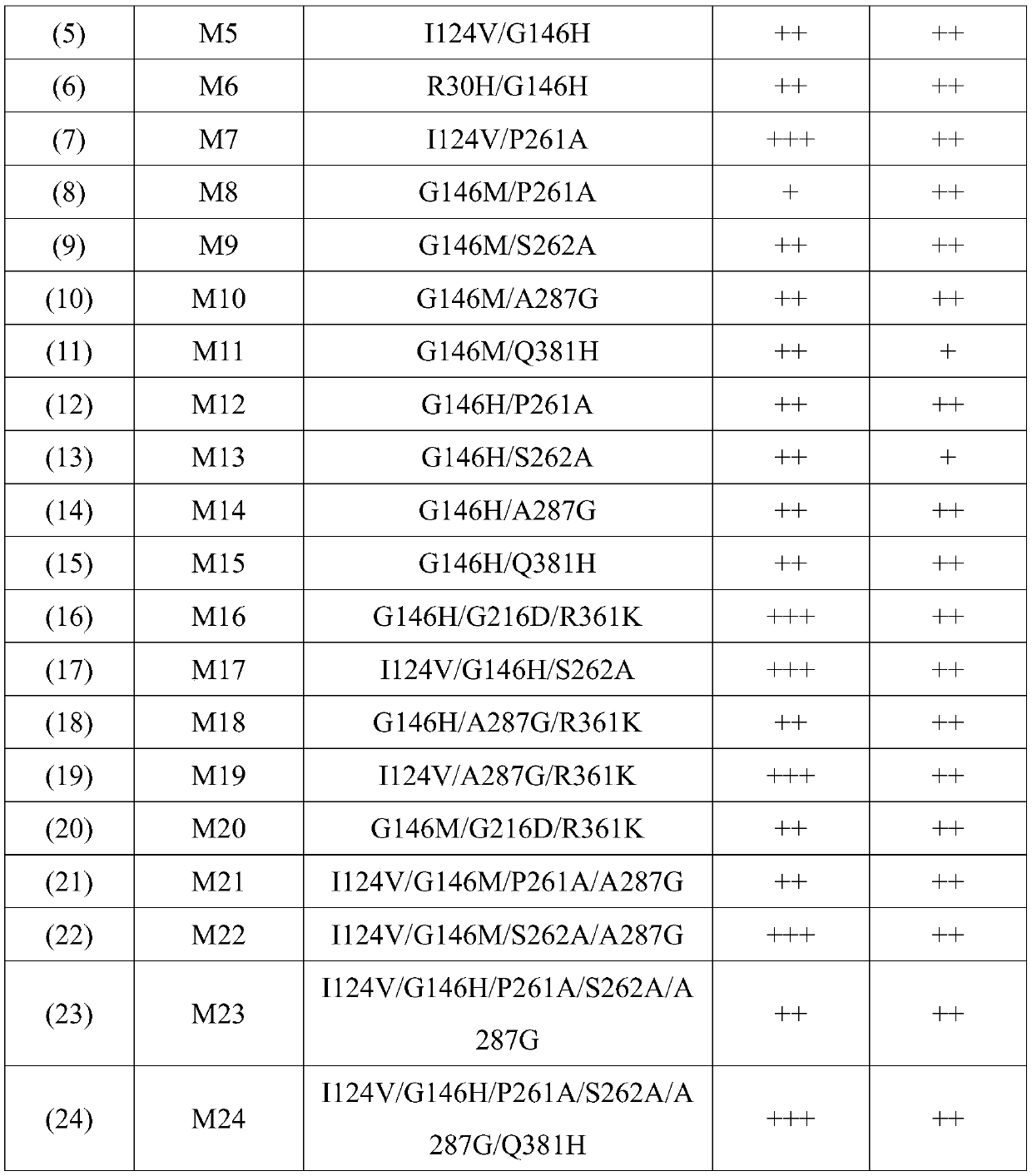
[0073] Example 2 Combinatorial mutation of BstFDH
[0074] On the basis of the mutations in Example 1, some mutation points were combined to obtain mutants with significantly improved affinity for coenzyme and substrate. The sequences of these mutants and the multiples of the increased affinity of these mutants for coenzyme and substrate are listed in in FIG. 1. In the list of Table 1, the sequence numbers respectively refer to a series of sequences corresponding to the back of Table 1; in the mutant affinity increase fold, a plus sign "+" indicates that the mutant protein ratio is shown by SEQ ID No. 2 in the sequence table. The affinity of the protein composed of the amino acid sequence to the substrate or coenzyme is increased by 0.1-1 times; the two plus signs "++" indicate that the mutant protein is compared to the protein composed of the amino acid sequence shown in SEQ ID No. 2 in the sequence table. The affinity of the substance or coenzyme is increased by 1-4 times; the...
Embodiment 2
[0103] Example 2 Expression and catalytic efficiency of recombinant BstFDH cat / K m Determination of
[0104] Inoculate recombinant E. coli BL21(DE3) / pET22b-BstFDH into 100mL LB medium containing 50μg / ml ampicillin, shake culture at 37℃ for 12 hours, and then inoculate 1%(v / v) Put the volume into a 500ml Erlenmeyer flask containing 100ml LB medium (containing 50μg / ml ampicillin), and place it on a shaker at 37°C and 180rpm for shaking culture. When the OD of the medium is 600 When it reached 0.6, IPTG with a final concentration of 0.2mmol / L was added as an inducer and induced at 16°C for 24h. The culture broth was centrifuged at 8000×g for 10 min, the cells were collected, and washed twice with saline to obtain resting cells. Suspend the cells obtained in 100ml of culture medium in 10ml of potassium phosphate buffer (100mM, pH 7.0), and sonicate them in an ice-water bath as follows: 400W power, working 4s, intermittent 6s, 99 cycles, 12000 at 4℃ After centrifugation at ×g for 4...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com