Polypeptide sequence specifically binding to avian bursal virus vp2 protein and its application
A specific and viral technology, applied in the field of molecular biology, can solve the problems of B-lymphocyte killing in the bursa of Fabricius, decreased immunity of chickens, vaccine immune failure and other problems, and achieve rapid and low-cost detection, rapid artificial synthesis, specific good sex effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Molecular docking and screening of virtual peptide library
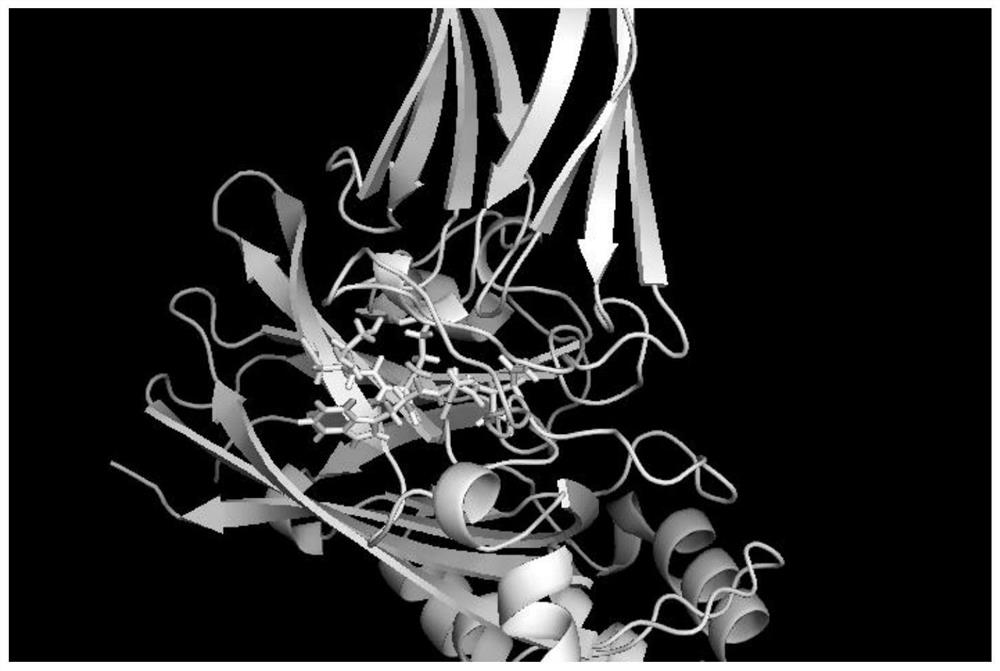
[0024] 1. Preparation of VP2 protein crystal structure
[0025] With the help of computer-aided design software, the amino acid sequence and crystal structure of IBDV VP2 protein were analyzed, and the 8th to 438th amino acid residues were selected as the docking region for molecular docking.
[0026] 2. Design of virtual peptide library
[0027] The present invention adopts the method of extending amino acid residues one by one. First, several amino acid libraries with the highest scores are docked with the structure of the target protein one by one. Until the best docking result is achieved. The peptide sequence generated by the virtual peptide library preferably has 2-9 amino acid residues.
[0028] 3. Evaluation of docking results
[0029] Using molecular docking software, comprehensively evaluate the free energy, hydrogen chain, van der Waals force and other mechanical parameters of peptide-...
Embodiment 2I
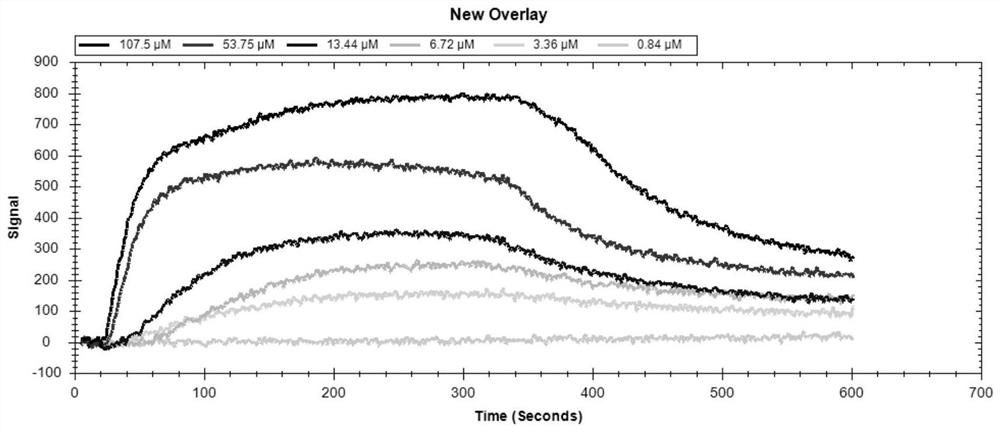
[0030] Example 2 Affinity Identification of IBDVL9-15 Sequence and Artificially Expressed VP2 Protein
[0031] 1. The first is the preparation of the VP2 protein chip, using the EDC / NHS method to covalently couple the IBDV VP2 protein (5 μg / mL) to the carboxyl chip of LSPR. When the LSPR Single≥500pm, it indicates that the VP2 protein coupling is successful. The sensor can be used to measure the interaction between IBDV VP2 protein and IBDV L9-15 sequence.
[0032] 2. The second is to run the sensor to obtain a stable signal baseline. Start running PBS buffer (pH 7.4) at the highest flow rate of 150 μL / min until the signal baseline of the sensor reaches a stable level. Then, reduce the flow rate to 20 μL / min and prepare for loading. Sample.
[0033] 3. Use PBS buffer to synthesize the solid phase, and prepare the dry powder of IBDVL9-15 modified by biotinylation at the amino terminal to the set concentration in advance, and then inject 300 μL of IBDVL9-15 solution into the s...
Embodiment 3I
[0035] Example 3 ELISA Identification of IBDVL9-15 Polypeptide and Artificially Expressed IBDV VP2 Protein
[0036] 1. Dilute the purified IBDV VP2 protein with CBS solution (pH9.6) to 10 μg / mL, and add it to an ELISA 96-well plate at a volume of 50 μL / well; express the purified protein from different viruses in the same way, that is, classical swine fever Viral E2 protein (CSFV-E2), porcine circovirus type 2 cap protein (PCV2-cap) and 1% bovine serum albumin (BSA), PBS buffer for ELISA 96-well plate coating, as a control, overnight at 4 °C quilt.
[0037] 2. Before blocking with 1% BSA solution, the ELISA plate was washed 5 times with PBST buffer. For each washing item, the reaction wells of the ELISA plate were filled with PBST, left to stand for 1 min, and then the PBST in the wells was discarded, and repeated 5 times. Subsequently, 1% BSA was added to the reaction wells of the ELSIA plate, placed in a 37° C. incubator, and left to stand for 1 h.
[0038] 3. Use PBS buffer...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com