Primer set for detecting trichina gene by loop-mediated constant temperature amplification, application and method
A loop-mediated isothermal, primer set technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial assay/inspection, etc. problems, to achieve the effect of easy promotion, improved detection sensitivity, and accurate detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0034] The specific detection of embodiment 2 primers
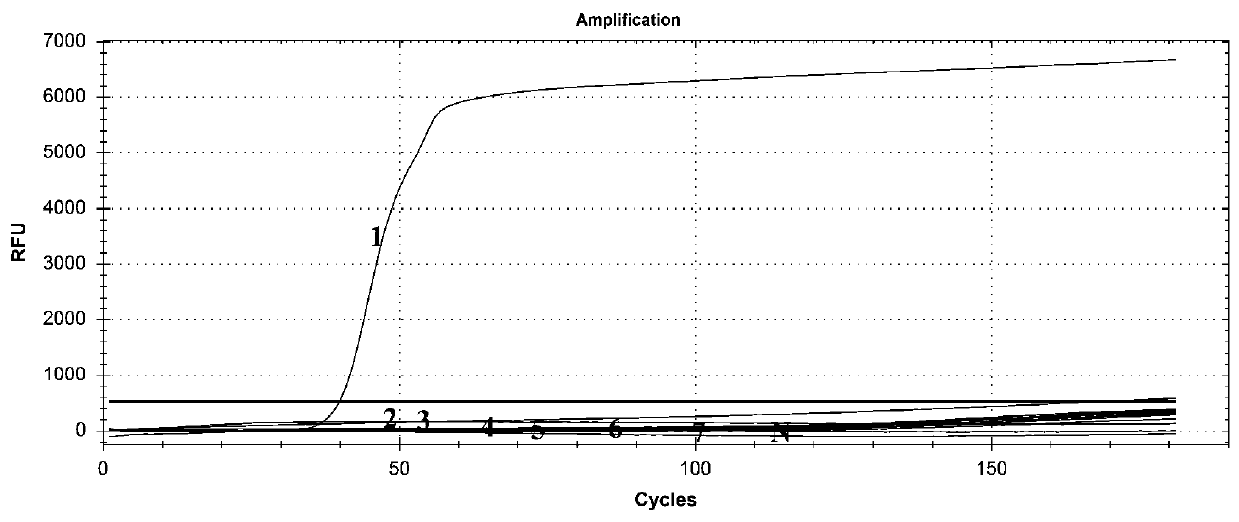
[0035] Using the genomic DNA of Trichinella spiralis, Angiostrongylus cantonensis, Anisakis elegans, gnathostomum, pinworm, Schistosoma japonicum and Trypanosomiasis as templates, add each template into a reaction tube, and add primer sets F3, B3, FIP, BIP , LB and LF, 8U Bst 2.0 DNA polymerase, 1.5mM dNTP, 7.5mM MgSO4 and ddH2O, in addition, add 1μl of SYTO13 fluorescent dye with a concentration of 25μM, and perform the reaction on Bio-Rad’s fluorescent quantitative PCR instrument. The specific reaction procedure For: react at 65°C for 1.5h and collect fluorescence data, react at 95°C for 5min.
[0036] The result is as figure 1 As shown, the primer set can detect the genomic DNA of Trichinella spiralis, but can not detect the genomic DNA of Angiostrongylus cantonensis, Anisakis elegans, gnathostomum, pinworm, schistosomiasis and Trypanosoma japonicum, indicating that the primers have good specificity .
Embodiment 3
[0037] Sensitivity detection of embodiment 3 primers
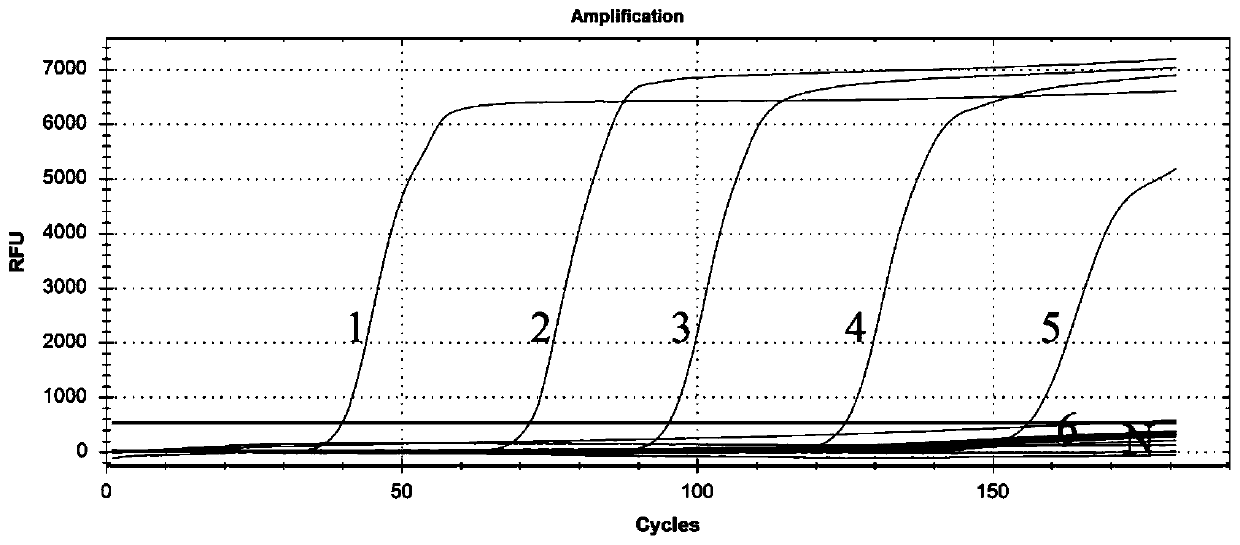
[0038] Dilute Trichinella spiralis genomic DNA to 17pg, 1.7pg, 170fg, 17fg, 1.7fg and 0.17fg and add them to reaction tubes respectively. Primers F3, B3, FIP, BIP, LB and LF, 8U Bst are also added to the tubes 2.0 DNA polymerase, 1.5mMdNTP, 7.5mM MgSO4 and ddH2O, in addition, add 1μl of SYTO13 fluorescent dye with a concentration of 25μM, and react on Bio-Rad’s fluorescent quantitative PCR instrument. The specific reaction program is: 65℃ for 1.5h and Fluorescence data were collected and reacted at 95°C for 5 minutes.
[0039] The result is as figure 2 As shown, the primer and reaction system can detect at least 1.7fg of Trichinella spiralis genomic DNA, and have good detection sensitivity.
Embodiment 4
[0040] The detection of embodiment 4 pyrophosphatase detection sensitivity
[0041] Trichinella spiralis genomic DNA was diluted to 17pg, 1.7pg, 170fg, 17fg, 1.7fg and 0.17fg and added to reaction tube 1 respectively. Primers F3, B3, FIP, BIP, LB and LF were also added to the tube, 8U Bst 2.0 DNA polymerase, 1.5mM dNTP, 7.5mM MgSO4 and ddH2O, in addition, add 8U pyrophosphatase, react in a 63°C water bath for 40 minutes, and then mix the reaction product with 4uL molybdic acid-tartaric acid in reaction tube 2 Potassium oxyantimony solution (21mM molybdic acid, 2mM potassium oxyantimony tartrate, 5M sulfuric acid) and 106uL distilled water, and 2uL 10% ascorbic acid and 106uL double distilled water solution in reaction tube 3 are fully mixed, if the solution turns blue, it is positive , which is negative if it remains colorless.
[0042] The result is as image 3 As shown, the pyrophosphatase detection system of the present invention can detect at least 1.7 fg of Trichinella ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com