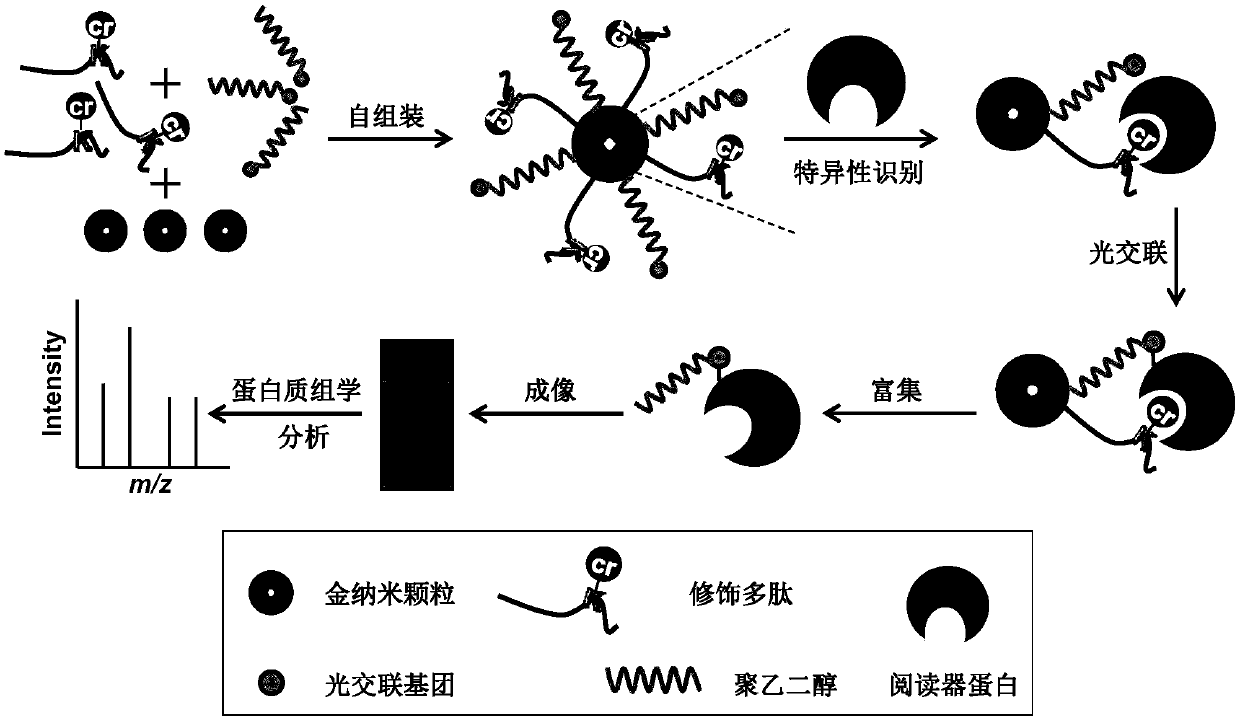
Polypeptide probe and application of polypeptide probe to identification of post-translational modification binding protein
A post-translational modification and protein-binding technology, which is applied in the direction of material inspection products, measuring devices, instruments, etc., can solve problems such as difficult identification and identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0111] The preparation of embodiment 1 polypeptide probe
[0112] The numbers and sequences of the five polypeptides are: ①: ARTKQTARKSC, ②: ARTK(me3)QTARKSC, ③: ARTKQTARKSTGGKAC, ④: ARTKQTARK(cr)STGGKAC, ⑤: ART(ph)K(me3)QTARKSC. The polypeptides were synthesized by Beijing Yaguang Biotechnology Co., Ltd. For the convenience of expression, the numbering of the polypeptide probes used in the following examples is consistent with the numbering of the polypeptides carried on it, and the above-mentioned corresponding relationship of numbering is used.
[0113] Mercapto polyethylene glycol coupled benzophenone, the molecular formula is HS-(CH 2 CH 2 O) 13 -C 6 h 4 COC 6 h 5 , synthesized by Shanghai Tuoyang Biotechnology Co., Ltd.
[0114] Gold nanoparticles (particle size 5nm) were purchased from BBI Company.
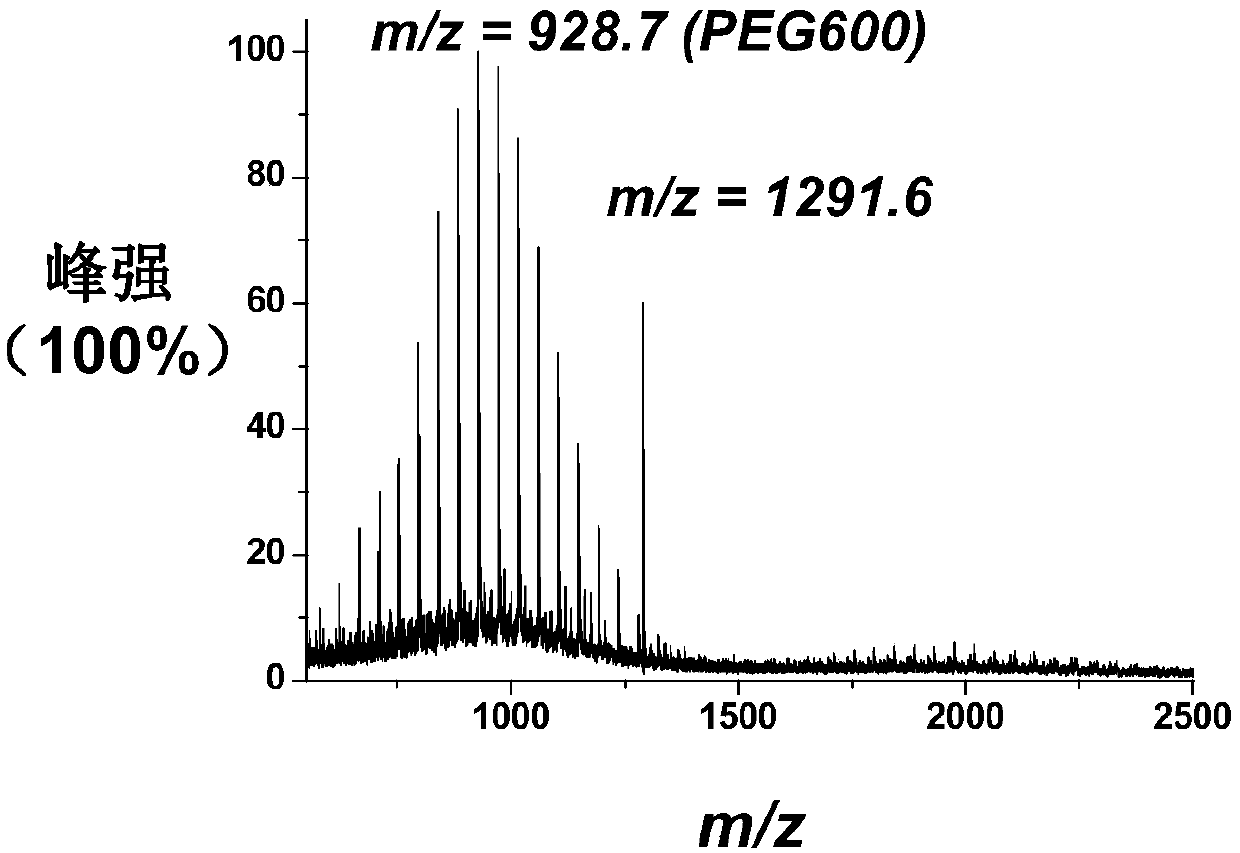
[0115] Preparation of polypeptide probe: Mix 2 μL (1 mM) polypeptide with 1 μL (1 mM) mercaptopolyethylene glycol-coupled benzophenone and add to 50 μL (80 nM) gol...
Embodiment 2
[0117] Example 2 Recognition and Efficient Enrichment of Target Proteins by Polypeptide Probes
[0118] Using BPTF (which has a PHD domain, which is known to be able to bind and modify H3K4me3) as the target protein, the verification of the effect of using the polypeptide probe in Example 1 was carried out. The BPTF domain protein is self-expressed with reference to the literature (Angew Chem Int EdEngl.2016Jul 4,55(28):7993-7), and the identified structure is shown in the appendix Figure 6 .
[0119] Experimental group: Incubate the target protein (10μg) and the peptide probe ② (4pmol) in binding buffer solution (50mM Tris-HCl, pH 7.8, 200mM sodium chloride, 2.5mM magnesium chloride and 2.5mM potassium chloride) overnight at 4°C . Then the sample tube was transferred to ice, and irradiated under a 365 nm ultraviolet lamp for 15 min to initiate a photocrosslinking reaction. Uncrosslinked proteins are then washed with binding buffer. The captured target protein was separat...
Embodiment 3
[0123] Example 3: Under the background condition of cell lysate or BSA, the selective enrichment of target protein by polypeptide probe.
[0124] Also using BPTF as the target protein, the effect of using the polypeptide probe ② in Example 1 was verified.
[0125] Experimental group: mix target protein (10μg) with cell lysate (50μg) (or bovine serum albumin (BSA), 50μg) in a certain ratio, and then mix with polypeptide probe (4pmol) in buffer solution (50mM Tris-HCl , pH 7.8, 200mM sodium chloride, 2.5mM magnesium chloride and 2.5mM potassium chloride) were incubated overnight at 4°C. Then the sample tube was transferred to ice, and irradiated under a 365 nm ultraviolet lamp for 15 min to initiate a photocrosslinking reaction. Next, the uncrosslinked protein was washed three times with PBS buffer solution (50 mM, pH 7.4) containing 4M urea. The captured protein was separated by centrifugation, and the target protein was added to the sample buffer (250mM Tris-HCl pH 6.8, 10% ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com