Detection primer and kit for MTB
A technology for detecting primers and kits, which is applied in the determination/inspection of microorganisms, microorganisms, and methods based on microorganisms, etc. It can solve the problems of different amplification efficiencies, inability of target DNA molecules, affecting PCR efficiency and measurement response, etc., to achieve high Specificity and accuracy, improved detection efficiency, and excellent amplification efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Primer sequences for detection of MTB amplification
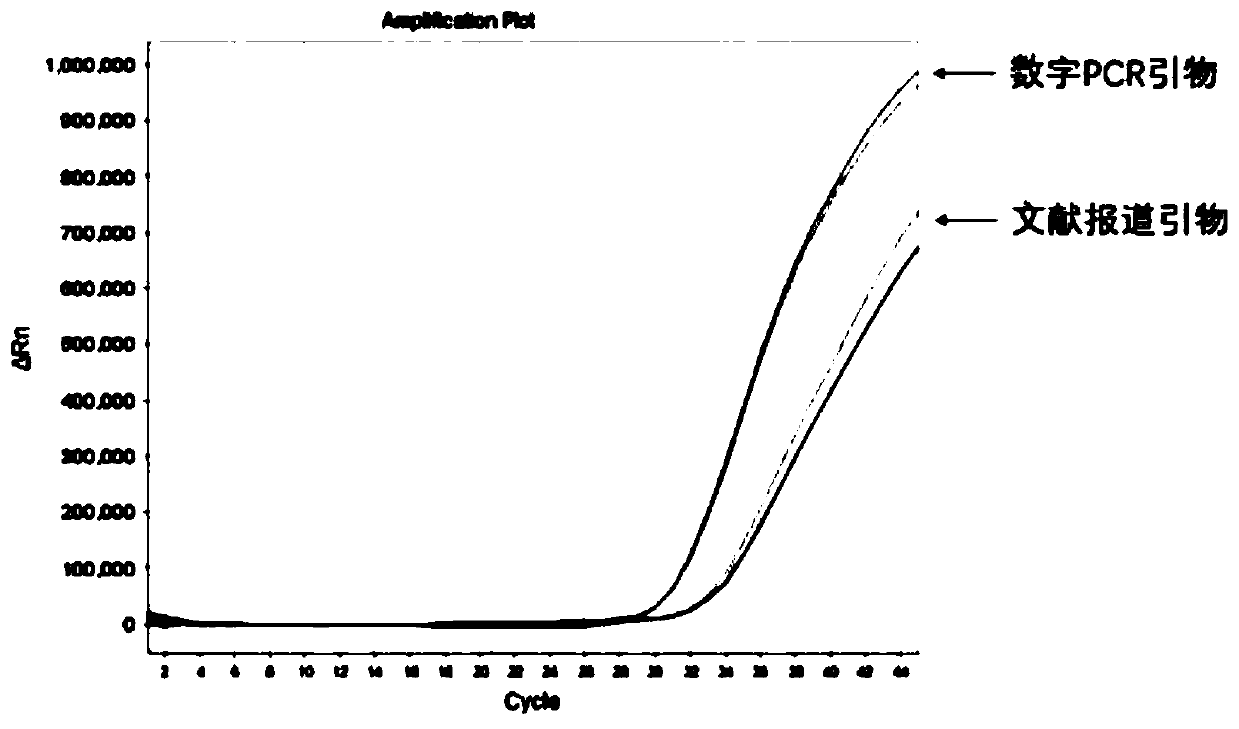
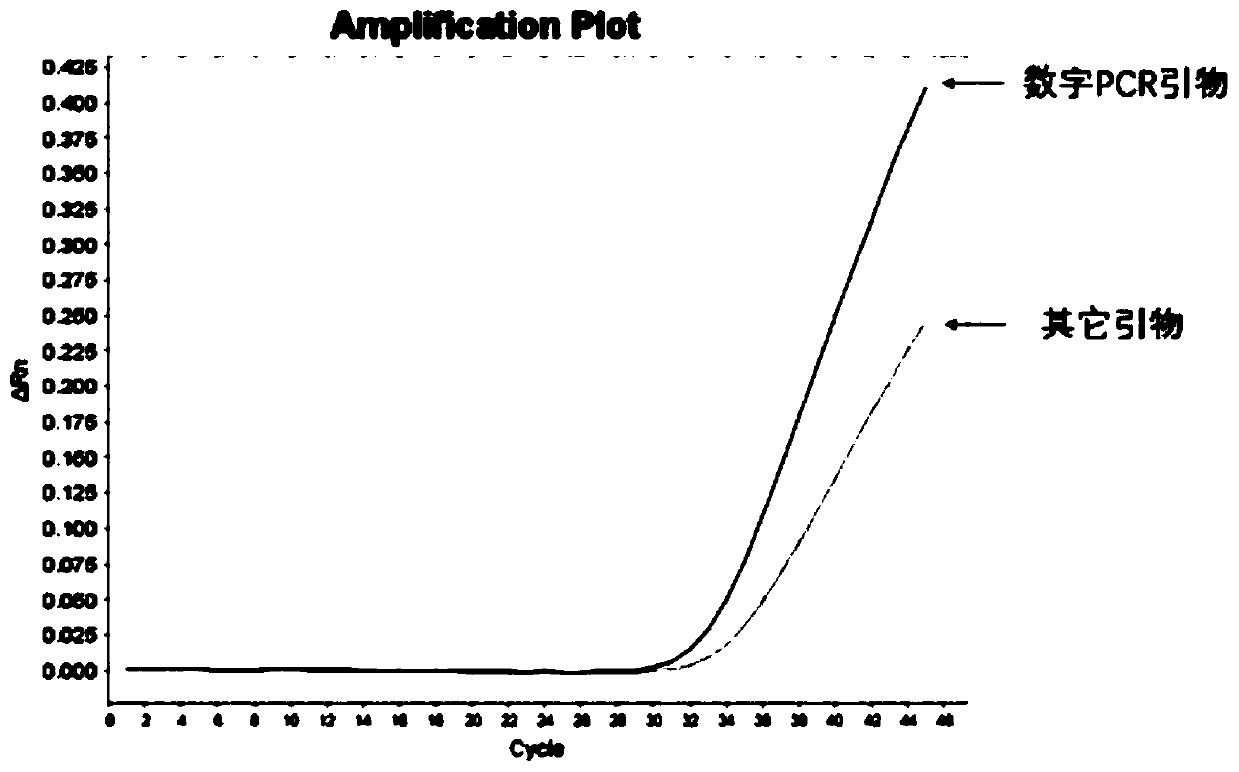
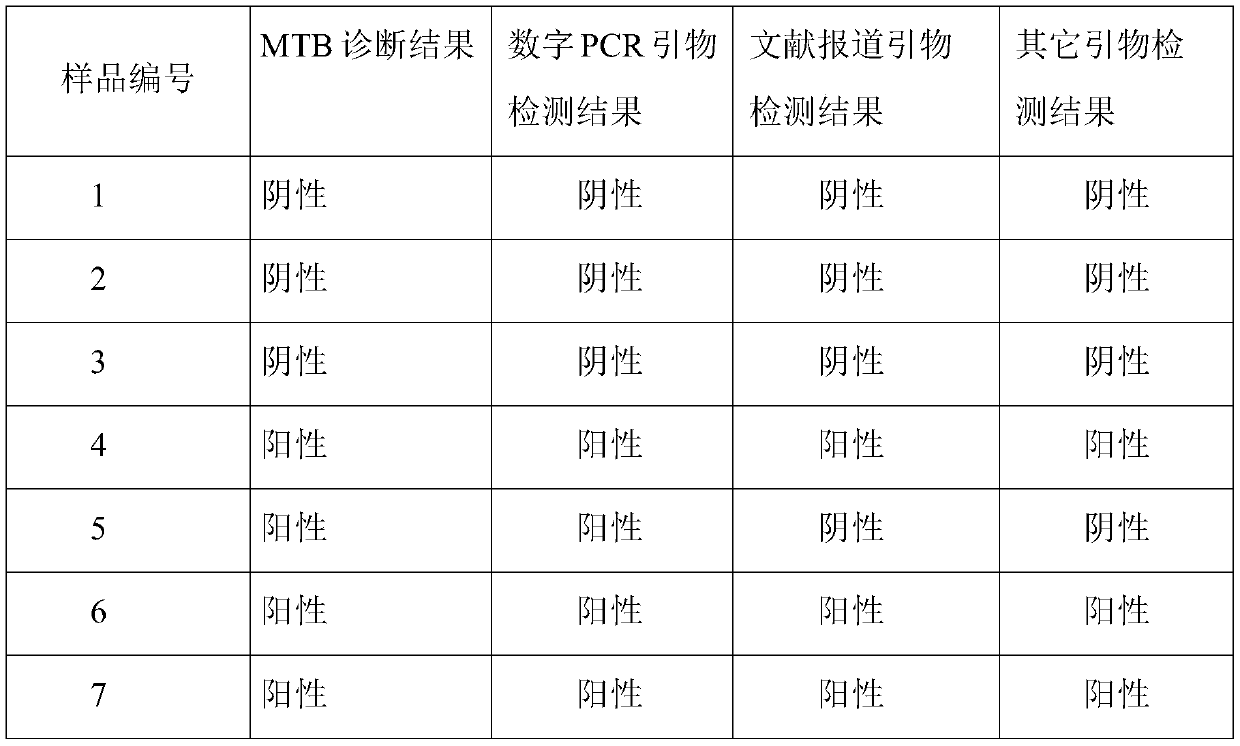
[0037] It includes the sequence shown in SEQ ID NO.: 1-3, and is the forward and reverse primers and MGB probe primers for amplifying MTB.
[0038]The base distribution of forward and reverse primers designed by the present invention is random, there is no continuous G or C base aggregation at the 3' end of the primer, and there is no self-complementary overlapping sequence at the 3' end, which can avoid the generation of hairpin structure and primer dimer . Related literature (Harkins K M, Buikstra J E, Campbell T, et al. Screening ancient tuberculosis with qPCR: challenges and opportunities [J]. Philosophical Transactions of the Royal Society B: Biological Sciences, 2015, 370(1660): 20130622.) reported primers As shown in SEQ ID NO.: 5-7, its reverse primer has a self-complementary sequence, which is easy to form a hairpin structure, which is not conducive to PCR amplification.
[0039] Forward primer reported in...
Embodiment 2
[0048] Testing MTB detection kits based on digital PCR technology
[0049] Including: Tissue DNA Extraction Kit (Extraction Kit from Capgemini, Cat. No. 158467); 70% Ethanol; Isopropanol; Chip Kit (Chip Kit from Hanghang Gene Company, Cat. No. V1); Detection System PCR Reaction Solution, positive control substance, negative control substance. The detection system PCR reaction solution includes: 2x Multiplex PCR Plus Buffer; dNTPMix; HotStar Taq Plus DNA Polymerase; forward and reverse primers and MGB probe primers for amplifying MTB shown in SEQ ID NO.: 1-3. In this article, 2x Multiplex PCR Plus Buffer, dNTP Mix; HotStar Taq PlusDNA Polymerase, and ROX dye are commercial routine reagents, and the corresponding brands can be directly retrieved. Experimenters can make corresponding commercial adjustments according to their own needs or self-preparation adjustments according to prescriptions Element.
Embodiment 3
[0051] A method for detecting MTB based on digital PCR technology
[0052] (1) Genomic DNA extraction from whole blood samples:
[0053] 1) Take 600 μL of blood and add 900 μl of red blood cell lysate, mix by inversion 10 times, place at room temperature for 1 min, during this period, invert and mix several times, centrifuge at 16,000 x g for 1 min, absorb the supernatant, leave the white blood cell precipitate, add 600 μl of cell lysate, Shake for 10s to thoroughly suspend and mix the white blood cells.
[0054] 2) Add 18 μl proteinase K solution, mix well, and incubate at 58 for 2 hours.
[0055] 3) Add 200 μl of protein precipitation solution, shake and mix at high speed for 20 seconds.
[0056] 4) Centrifuge at 16000x g for 1 min, leave the supernatant, and discard the precipitate.
[0057] 5) Add the supernatant from the previous step to a 1.5ml centrifuge tube filled with 600μl isopropanol in advance, and gently invert up and down for 50 times to mix well. At this tim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com