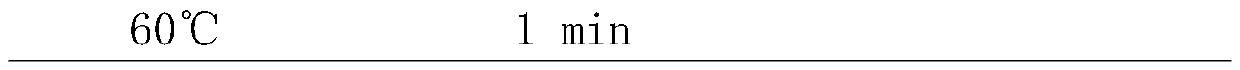Use method for de-host extraction test kit of a metagenome sample
A metagenomic and kit technology, applied in the field of dehosting extraction kits for metagenomic samples, can solve problems such as differences in susceptibility of wall-breaking lysis methods, host DNA residues, and deviations in the detection of representative microbial components of samples, and improve the Effective microbial sequencing data, short extraction process time, and good clinical application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
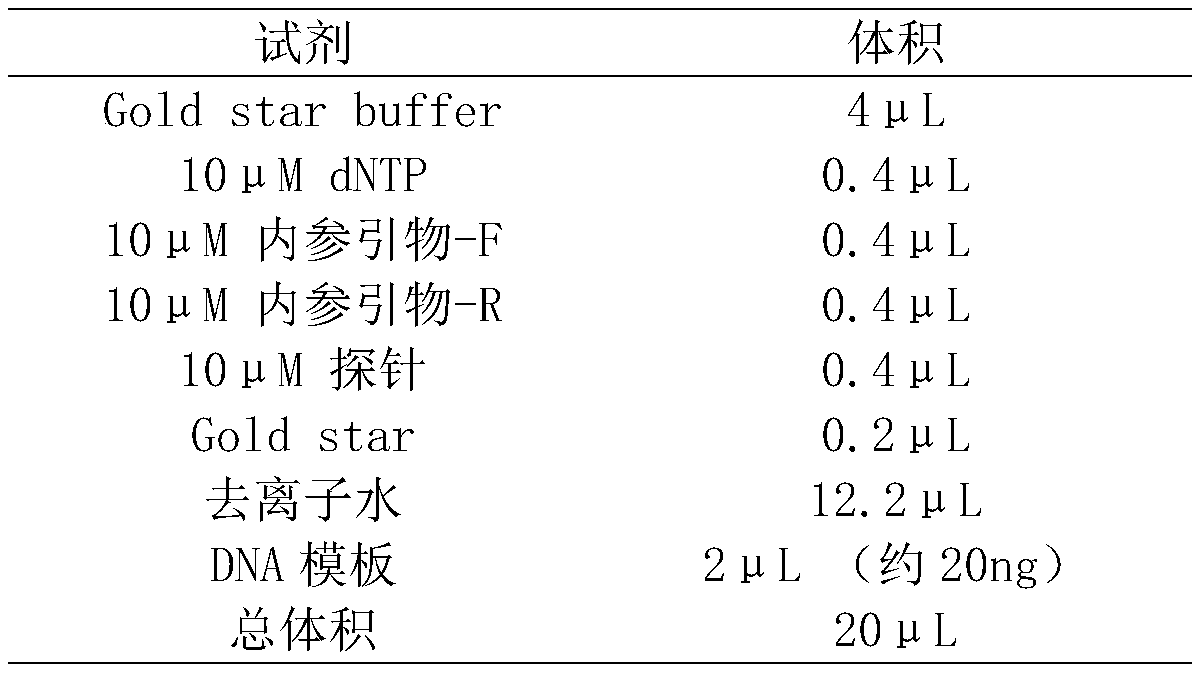
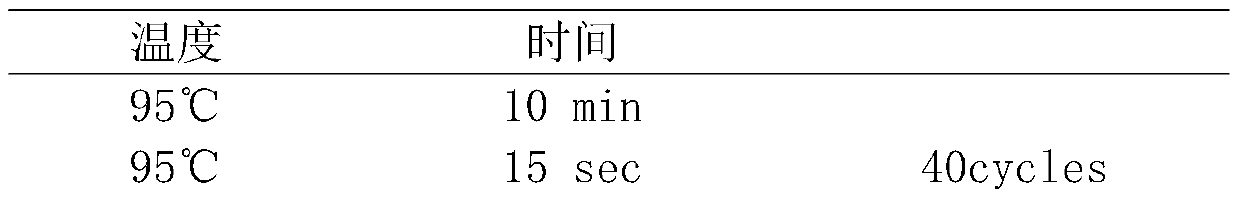
[0021] Sample handling:
[0022] (1) Simulated mixed sample: Take 5 μL each of Escherichia coli, Staphylococcus aureus, Enterococcus faecium, and yeast cultured overnight, and add it to 500 μL fresh blood sample (preserved in EDTA preservation tube) as a simulated sample.
[0023] (2) Clinical blood samples: 500 μL of blood samples from five patients with clinical infectious diseases were collected in 1.5 mL centrifuge tubes.
[0024](3) Preparation of trypsin digestion solution: 4g / L trypsin (1:2500), 30mM CaCl2, 100mM NaCl, 2mMKCl, 8mM Na2HPO4, 4mM KH2PO4, adjust the pH value to pH 8.0 with NaOH solution, and use a 0.45μM filter Sterilize and insoluble matter by filtration, store at 2-8°C for later use.
[0025] Clinical sputum samples: Take 500 μL of sputum samples from 5 patients with clinical infectious diseases in 1.5 mL centrifuge tubes, add 750 μL of trypsin digestion solution, and incubate at 37°C for 15-30 minutes to completely liquefy the samples.
[0026] A metho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com