Chitin deacetylase, encoding gene and application
A deacetylase, encoding gene technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve the problems of difficult separation, harsh reaction conditions, serious environmental pollution and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
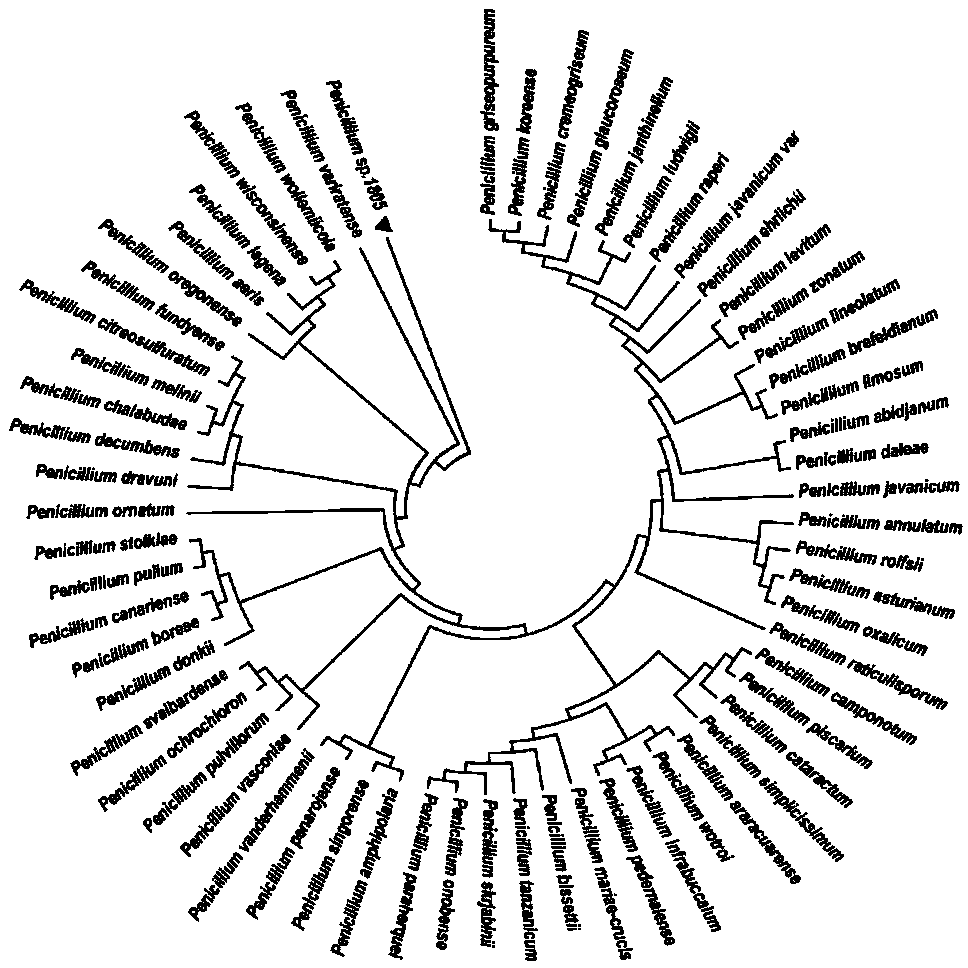
[0023] Embodiment 1: Phylogenetic tree construction
[0024] Penicillium (American Strain Classification Number Penicillium oxalicum 114-2, Taxonomy ID: 933388) Penicillium sp.1805 Isolation and Purification Fungal Genome Extraction Kit (Manufacturer: Sangon, Model: B518229-0050), Genome Extraction, and ITS1 and ITS4 The primers were cloned, the fungi were identified, and the sequencing results were compared on NCBI, and the sequences with close relatives were downloaded to construct the phylogenetic tree through MEGA7.
Embodiment 2
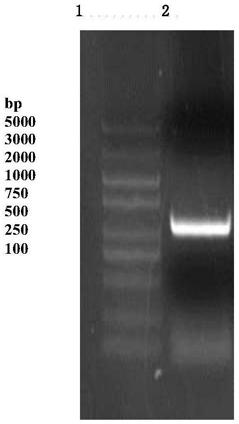
[0025] Example 2: Full-length cloning of chitin deacetylase gene.
[0026] RNA was extracted from Penicillium oxalicum (Penicillium oxalicum 114-2, Taxonomy ID: 933388), reverse-transcribed to form cDNA, and primers CDA-F: 5'-CTAGTCTAGAATGGCCCCCAAGAGAGTATTG-3'; CDA-R5'-CCGCTCGAGCTACTTCTTGAGCATCTGACC- 3'. The reverse transcribed cDNA was used as a template for PCR amplification. The PCR reaction conditions were: 94°C for 5min, 1 cycle; 94°C for 30s, 45°C for 30s, 72°C for 1min30s, 30 cycles; 72°C for 10min, 1 cycle. After the PCR product was analyzed by agarose gel electrophoresis, the target fragment was recovered by cutting the gel, connected to the pMD19-T vector and then sequenced.
[0027] Sequence listing:
[0028]Information on SEQ ID No.1
[0029] (a) Sequential features
[0030] Length: 927 nucleotides
[0031] Type: Nucleotide
[0032] Chain type: single chain
[0033] (b) Molecule type: DNA
[0034] Sequence description: The gene encoding chitin deacetylase ...
Embodiment 3
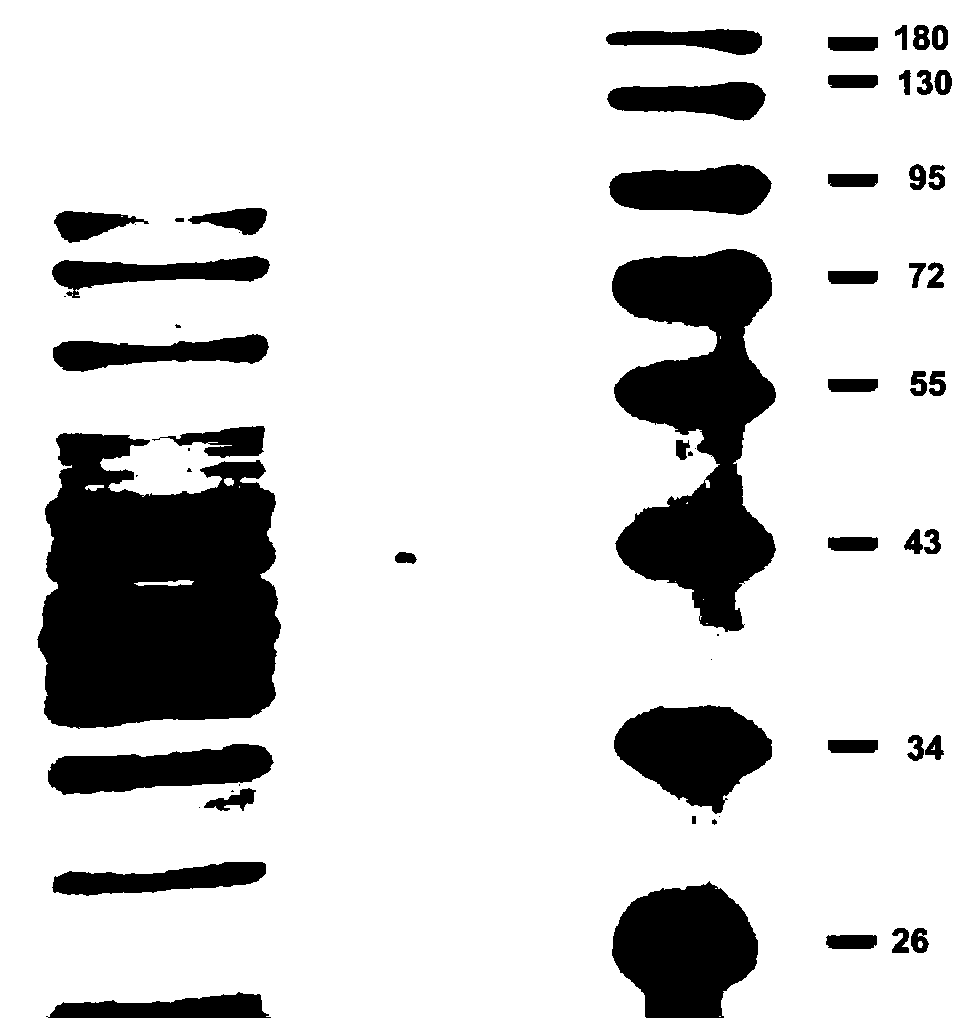
[0046] Embodiment 3: the heterologous expression of target gene
[0047] The PCR amplified product and the pET-22b vector plasmid were double-digested with BamHI and HindIII restriction endonucleases, digested at 37°C for 12h, and the fragment was ligated to the pET-22b vector plasmid, and ligated at 16°C for 18h. Take 5 μL of the ligation product and transfer it into E.coliDH5α, select a single clone, and perform bacterial liquid PCR detection and sequencing verification to obtain the recombinant plasmid psccda-PET-22b.
[0048] The recombinant plasmid extracted and sequenced correctly was transferred into Escherichia coli BL21 (DE3) expression-competent cells, and monoclonal PCR was selected for verification. The correct single clone was cultured in 10 mL LB medium containing ampicillin with a final concentration of 100 μg / ml for 12 hours as the seed solution. The seed solution was added to 1L fermentation medium at a volume ratio of 1:100, induced for 12 hours at 16°C, cen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com