Fast nucleic acid extraction, sequencing and identification method based on bacterial 16S rDNA sequence
An identification method and nucleic acid technology, which are applied in the field of rapid nucleic acid extraction and sequencing identification based on bacterial 16S rDNA sequences, can solve the problems of long extraction time, inconvenient steps, limited application scope, etc., and achieve the effect of longer solution time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] DNA extraction from samples
[0062] The sample is the plate streak bacteria or culture of the bacterial strain to be identified;
[0063] Add the plate streaked bacteria or culture into the test tubes, add 20uL cell lysate to each test tube, shake and mix well, let stand at room temperature for 10min-30min, then dilute 10-50 times, shake and mix again, and then high-speed Centrifuge for 2 minutes, and take the centrifuged supernatant as the DNA template to be detected.
[0064] In addition, the cell lysate is a mixture of sodium hydroxide solution and sodium dodecyl sulfate solution, wherein the mass concentration of sodium hydroxide solution is 0.1M-0.5M, wherein the mass percentage of sodium lauryl sulfate solution The concentration is 1%-3%.
Embodiment 2
[0066] PCR amplification reaction
[0067] The general primers are as follows:
[0068] The upstream primer 27F of the universal primer is shown in SEQ ID No.1,
[0069] The downstream primer 1492R of the universal primer is shown in SEQ ID No.2.
[0070] The reaction system of the PCR amplification reaction is calculated as 30uL:
[0071]
[0072] The amplification program of the PCR amplification reaction is:
[0073]
[0074] Repeat for 25 cycles;
[0075] Extend at 72°C for 7 minutes;
[0076] Stop the reaction at 4°C and store.
[0077] It should be noted that the sequences of the universal primers are as follows:
[0078] Forward primer 27F (SEQ ID No.1): 5'-AGAGTTTGATCCTGGCTCAG-3',
[0079] Reverse primer 1492R (SEQ ID No. 2): 5'-TACGGCTACCTTGTTACGACTT-3'.
Embodiment 3
[0081] Analysis of PCR amplification products
[0082] The specific method is as follows:
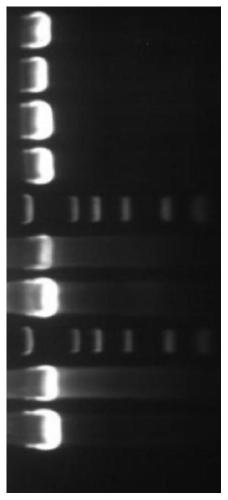
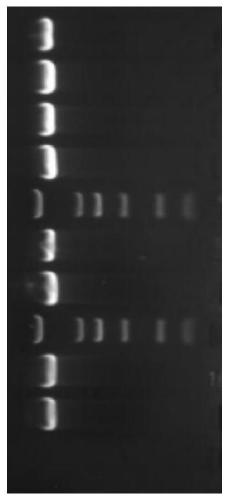
[0083] (A) Take PCR amplification product, prepare agarose gel and carry out electrophoresis, the voltage of electrophoresis is 100V, after electrophoresis finishes, use gel imager to observe, take pictures and record experimental results;
[0084] (B) cutting out the nucleic acid amplification product of the agarose gel after electrophoresis in step (A), extracting and purifying the nucleic acid;
[0085] (C) The nucleic acid purified in step (B) is used to obtain a spliced sequence using a general-purpose sequence splicing software, and then the spliced sequence is subjected to PCR amplification reaction, and the product of the PCR amplification reaction is sequenced and sequence spliced to obtain 16S rDNA sequence;
[0086] (D) importing the DNA sequence of the corresponding fragment in step (C) into a standard nucleic acid sequence database for sequence comparison to identif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com