Recombinant saccharomycetes for producing ursolic acid and oleanolic acid and construction method and application of recombinant saccharomycetes
A technology of recombinant bacteria and homologous recombination, applied in the biological field, can solve the problems of low yields of ursolic acid and oleanolic acid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1, the construction of synthetic α-amyresinol and β-amyresinol Saccharomyces cerevisiae recombinant bacteria
[0036] 1. Module construction
[0037] According to the gene sequence of the multifunctional balsamol synthase CrAS (Catharanthus roseus, GenBank: JN991165) in periwinkle, the codon optimization for Saccharomyces cerevisiae was obtained by chemical synthesis (Nanjing GenScript Biotechnology Co., Ltd.) Gene CrAS (SEQ ID NO.1) encoding the multifunctional balsamol synthase.
[0038] Squalene epoxidase coding gene ERG1 (SEQ ID NO.2), promoter P GAL1 (SEQ ID NO.8), P TEF1 (SEQID NO.9) and terminator T CYC1 (SEQ ID NO. 10), T ADH1 (SEQ ID NO.11) are all from Saccharomyces cerevisiae W303-1a genome; Selection marker gene HIS3 is from plasmid pxp320 (purchased from Addgene, Inc. www.addgene.org ).
[0039] Using the genome of Saccharomyces cerevisiae W303-1a (USA, ATCC: 208352) as a template, P GAL1 -CrAS-F (SEQ ID NO.12) and P GAL1 -CrAS-R (SEQ ID...
Embodiment 2
[0048] Embodiment 2, the construction of Saccharomyces cerevisiae strain Sc310LCZ02
[0049] Saccharomyces cerevisiae endogenous tHMG1 (SEQ ID NO.3), ERG20 (SEQ ID NO.4), and promoter P PGK1 (SEQ ID NO. 30), P TDH3 (SEQ ID NO.31) and terminator T CYC1 (SEQ ID NO.10) are all from the Saccharomyces cerevisiae W303-1a genome. PRS304 plasmid was purchased from BioVector (http: / / www.biovector.net) and PRS405 plasmid was purchased from Biofeng (www.biofeng.com).
[0050] 1. Construction of plasmid ptHMG1
[0051] Using the genome of Saccharomyces cerevisiae W303-1a as a template, the primers P TDH3 -F-ApaI (SEQ ID NO.32), P TDH3 -R-tHMG1 (SEQ ID NO.33) and tHMG1-T CYC1 -F (SEQ ID NO. 34), T CYC1 -R-PstI (SEQ ID NO.35) is primer, amplifies P TDH3 (SEQ ID NO.31) promoter and T CYC1 (SEQ ID NO.10) terminator, with P TDH3 -tHMG1-F (SEQ ID NO.36) and P TDH3 -R-tHMG1 (SEQ ID NO.37) is a primer for amplifying the tHMG1 gene (SEQ ID NO.3). By the fusion PCR condition fusion P in...
Embodiment 3
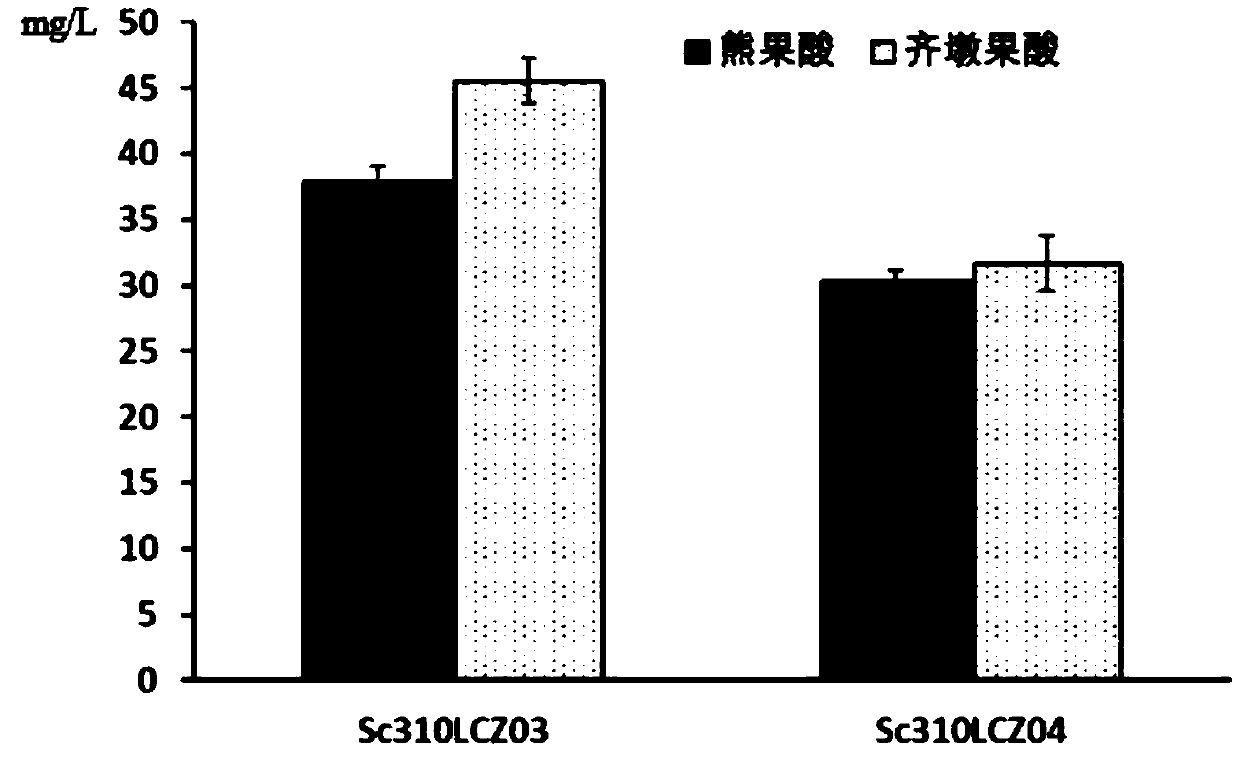
[0055] Embodiment 3, the construction of synthetic ursolic acid and oleanolic acid Saccharomyces cerevisiae recombinant bacteria
[0056] According to vinca C-28 oxidase CYP716AL1 (Catharanthus roseus, GenBank: JN565975), C-28 oxidase CYP716A12 (Medicagotruncatula, GenBank: ABC59076.1) and Arabidopsis The gene sequence of cytochrome-NADPH-reductase 1 gene AtCPR1 (Arabidopsis thaliana, GenBank: BT008426.1) in mustard was optimized for Saccharomyces cerevisiae, and then chemically synthesized (Nanjing KingScript Synthesis) to obtain the coding gene CYP716AL1 (SEQ ID NO.7) encoding the C-28 oxidase of vinca argentinol and CYP716A12 (SEQ ID NO.5) and Arabidopsis cytochrome-NADPH-reductase 1 encoding gene AtCPR1 (SEQ ID NO.6) in Arabidopsis thaliana. promoter P TDH3 (SEQ ID NO. 31), P PGK1 (SEQ ID NO.30) and terminator T CYC1 (SEQ ID NO. 10), T ADH3 (SEQ ID NO.44) are all from Saccharomyces cerevisiae w303-1a genome; the selection marker gene ura3 is from plasmid pxp218 (Addge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com