SUMOylation peptide fragment enrichment method based on anion exchange chromatographic column
A technology of exchange chromatography and anion, applied in the field of enrichment of SUMOylated peptides, can solve the problem of inability to accurately reflect the real modification state of proteins, and achieve the effect of improving identification coverage, high selectivity and high enrichment selectivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
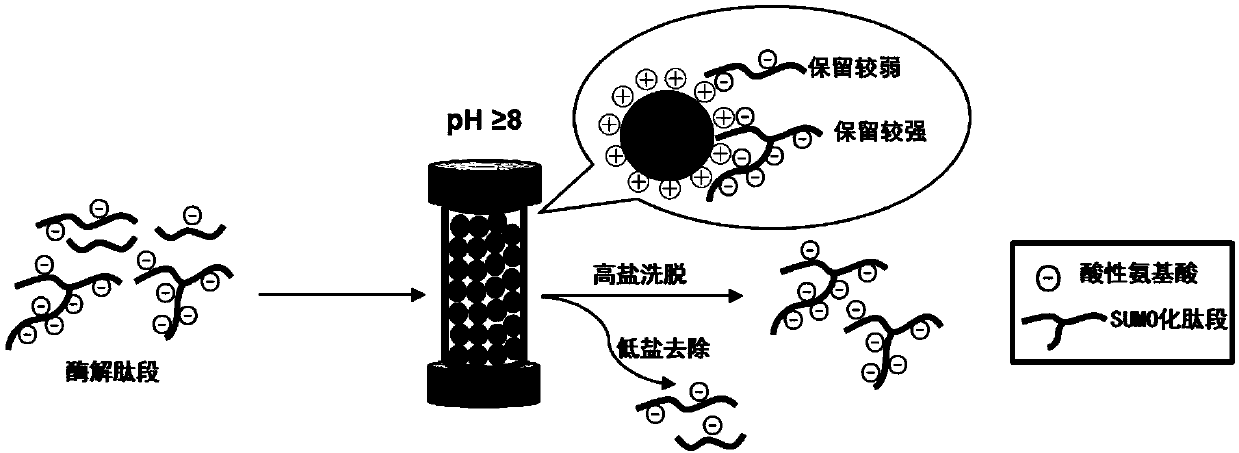
[0029] Such as figure 1 As shown, the basic site of the protein is first digested, and the SUMOylated peptide after digestion has multiple acidic amino acids, and the retention in anion exchange chromatography in an alkaline environment is stronger than that of the non-SUMOylated peptide. Anion-exchange chromatography was used to elute the weakly retained enzyme cleavage products first, and finally the peptide fractions with strong retention were collected, which were the enriched SUMOylated peptides.
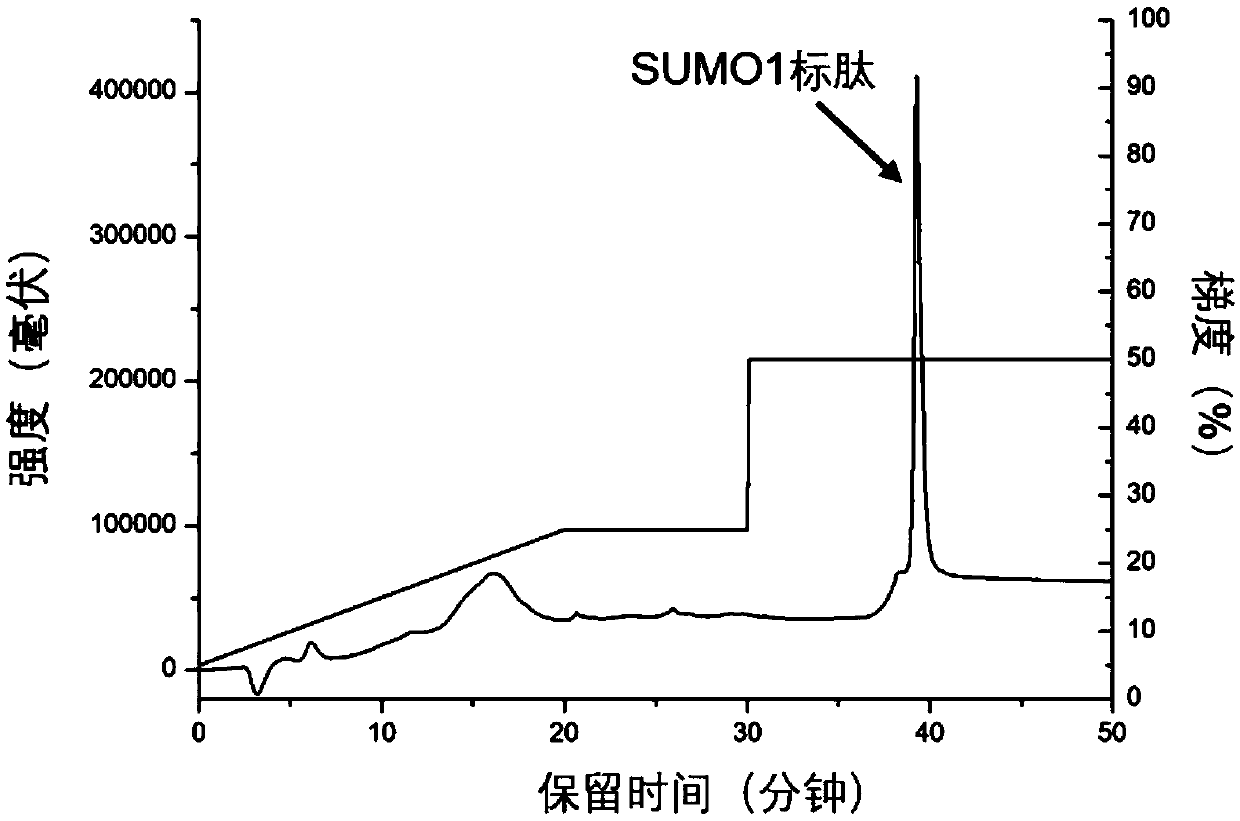
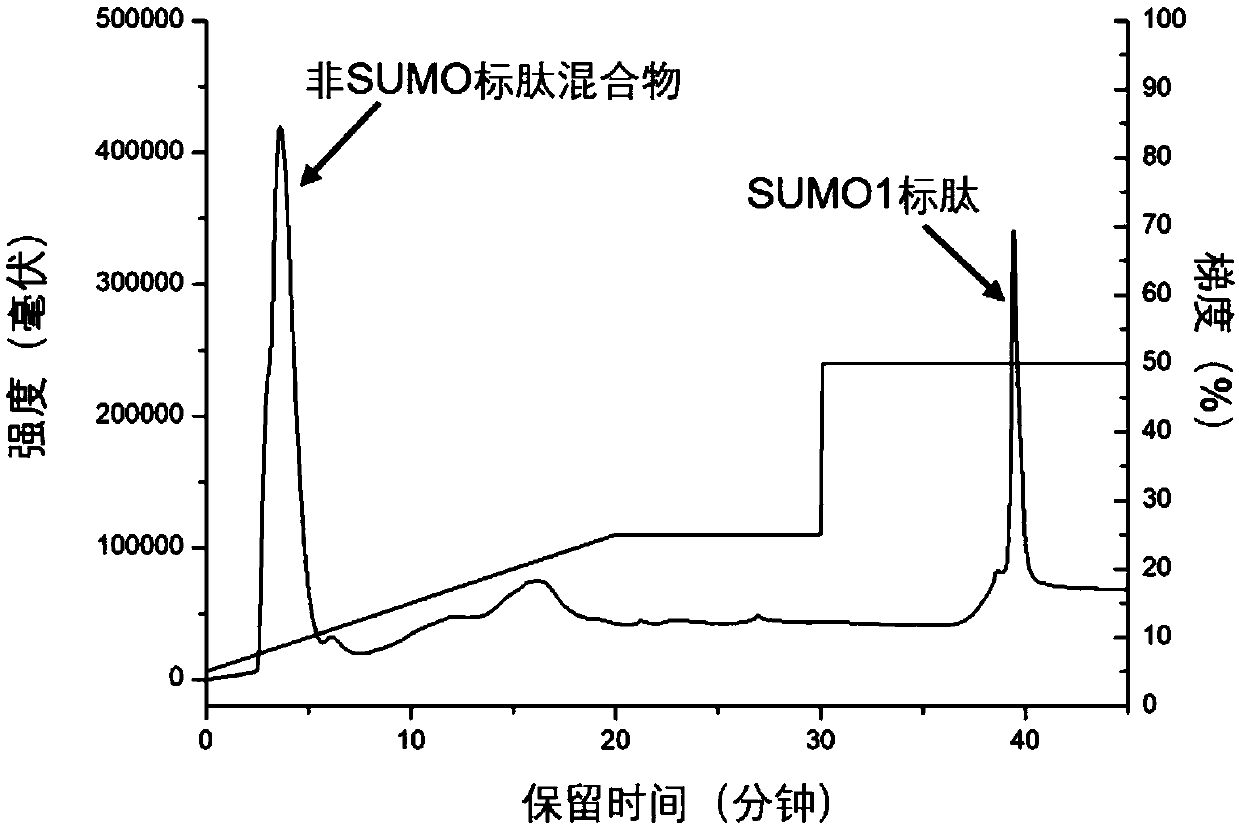
[0030] Take HeLa cells as samples, dissolve 100 μg of extracted protein in 100 μl of 8M urea in 50 mM ammonium bicarbonate buffer (pH 8), add 10 μl of 100 mM dithiothreitol, denature at 56°C for 1 hour, then add 10 μl of 300 mM iodoacetamide to react After 0.5h, add another 30μl of 100mM dithiothreitol, incubate for 10min, digest with trypsin, wherein the amount of enzyme is 1 / 10 of the sample mass, the temperature is 37℃, after 60min of enzymolysis, desalt, freeze-dry , redis...
Embodiment 2
[0032] Take HeLa cells as samples, dissolve 100 μg of extracted protein in 100 μl of 8M urea in 50 mM ammonium bicarbonate buffer (pH 8), add 10 μl of 100 mM dithiothreitol, denature at 56°C for 1 hour, then add 10 μl of 300 mM iodoacetamide to react After 0.5h, another 30μl of 100mM dithiothreitol was added, and after incubation for 10min, it was digested with Lys-C, wherein the enzyme dosage was 1 / 10 of the sample mass, and the temperature was 37°C. Dried, redissolved in 0.1% formic acid, and analyzed by mass spectrometry, the lysine at the carboxy-terminal of the peptide was cleaved efficiently and selectively.
Embodiment 3
[0034] Take HeLa cells as samples, dissolve 100 μg of extracted protein in 100 μl of 8M urea in 50 mM ammonium bicarbonate buffer (pH 8), add 10 μl of 100 mM dithiothreitol, denature at 56°C for 1 hour, then add 10 μl of 300 mM iodoacetamide to react After 0.5h, add another 30μl 100mM dithiothreitol, incubate for 10min, and use Arg-C to digest, wherein the amount of enzyme is 1 / 10 of the sample mass, and the temperature is 37℃. After 60min of enzymatic hydrolysis, desalt, freeze Dried, redissolved in 0.1% formic acid, and analyzed by mass spectrometry, the arginine at the carboxy-terminal of the peptide was cleaved efficiently and selectively.
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com