Primer for amplifying novel coronavirus and application of primer
A coronavirus, a new type of technology, applied in biochemical equipment and methods, microbial determination/inspection, resistance to vector-borne diseases, etc., can solve the problem of no novel coronavirus nucleic acid loop-mediated isothermal amplification primer detection technology, etc. , to achieve the effect that is easy to obtain, highly conservative, and easy to quantify
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 Preparation of a kit for detecting novel coronavirus
[0056] Step 1: Prepare Positive Control Standard
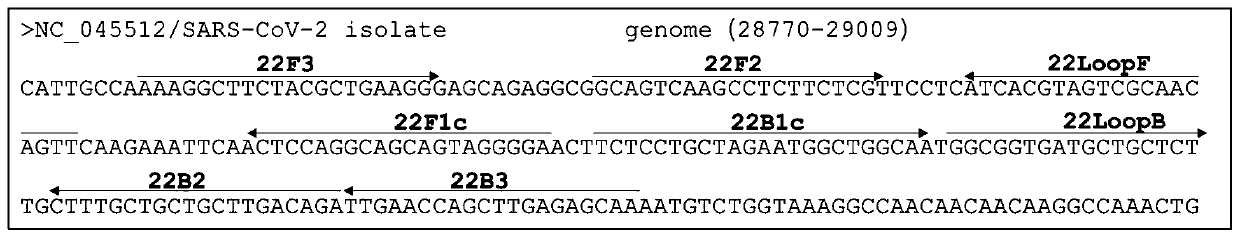
[0057] Entrusted Nanjing GenScript Biotechnology Co., Ltd. to synthesize the N sequence of the nucleocapsid encoding gene of the new coronavirus reference strain (GenBank accession number: NC_045512.2, sequence from 28274 to 29533 in the genome). KpnI and BamHI restriction enzyme sites were introduced into the 5'-end and 3'-end of the gene respectively, and it was named SARS-CoV-2N gene.
[0058] Synthetic SARS-CoV-2N gene and The -3Z (Promega) plasmid vector was double digested with Kpn I and BamHI to obtain a linear vector with cohesive ends and an exogenous gene. The specific enzyme digestion reaction system is as follows:
[0059]
[0060]The enzyme digestion reaction was carried out at 37°C for 30 minutes. After the reaction, the linearized plasmid vector and the foreign gene were recovered with the Cycle-Pure PCR Product Recovery Kit (Omega Bi...
Embodiment 2
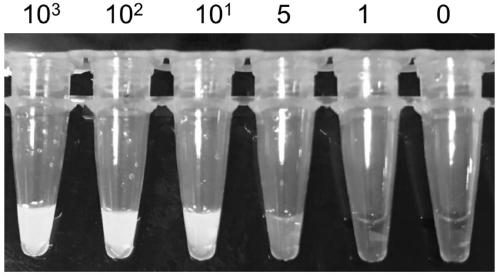
[0080] Verify the sensitivity of the detection results of the novel coronavirus loop-mediated isothermal amplification (RT-LAMP) using the primers described in Example 1 of the present invention. Include the following steps:
[0081] Dilute the RNA template: take the RNA prepared by in vitro transcription as the template (i.e. the positive control standard prepared in step 1 of the embodiment), and after quantification, dilute the RNA copy number concentration to 10 with sterile enzyme-free deionized water. 3 copies / μL, 10 2 copies / μL, 10 1 copies / μL, 5copies / μL and 10 0 copy / μL, and take the negative control as 0copy / μL template RNA.
[0082] Establishment of detection system
[0083] In a 0.2ml micro-reaction tube (Haijibio), add the reagents in the doses described in the table below to establish a loop-mediated isothermal amplification reaction system:
[0084]
[0085] After the reaction system is prepared, gently mix the liquid in the micro reaction tube, and c...
Embodiment 3
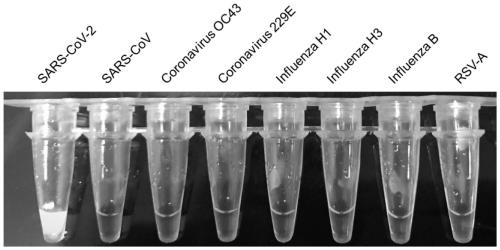
[0089] Verify the specificity of the detection results of the novel coronavirus loop-mediated isothermal amplification (RT-LAMP) using the primers described in Example 1 of the present invention. Including the following steps: Select the inactivated severe respiratory syndrome virus (SARS-CoV), coronavirus 229E, coronavirus OC43, H1 subtype influenza A virus (Influenza H1) in the NATtrol Respiratory Validation Panel 3 kit (ZeptoMetrix) Influenza H3, Influenza B, and Respiratory Syncytial Virus A (RSV-A) are 7 common respiratory virus controls for specific detection of the tested virus. Use the virus DNA / RNA extraction kit (Novizem Biotechnology Co., Ltd.) to extract the RNA of the above viruses, and use it together with the SARS-CoV-2 N gene transcript prepared in this study as the sample to be tested, and establish a loop-mediated isothermal amplification method. Increased response. React in a water bath at 60°C for 30 minutes. After the reaction is over, judge the reaction ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com