SNP markers related to assessment of incidence risks of NK/T cell lymphoma and application of SNP markers
A lymphoma and marker technology, applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems that the susceptibility of the population has not been verified, and the risk of NK/T cell lymphoma is low , to achieve the effects of convenient material collection, increased sensitivity, and improved applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
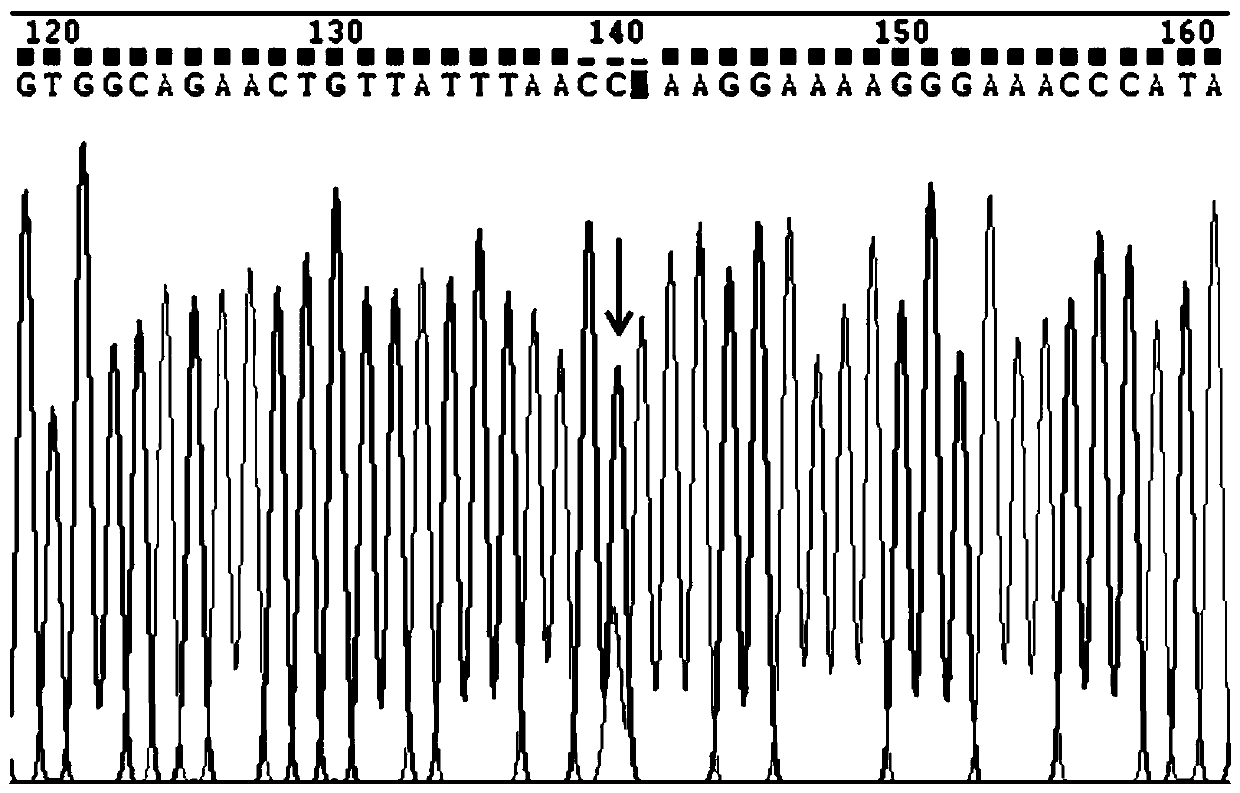
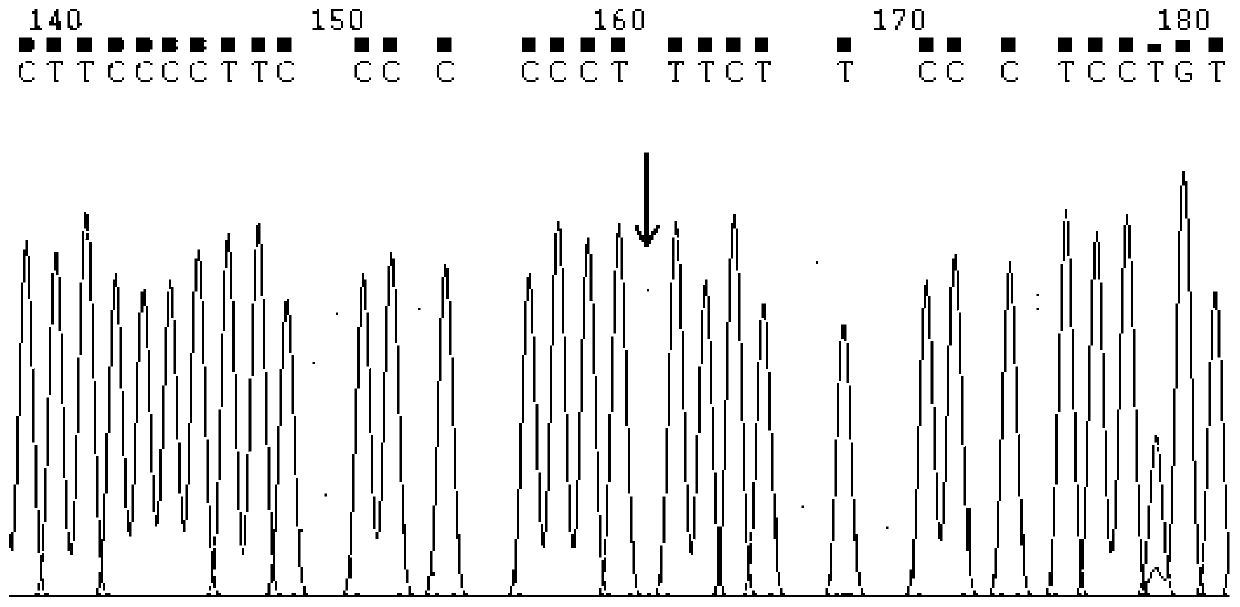
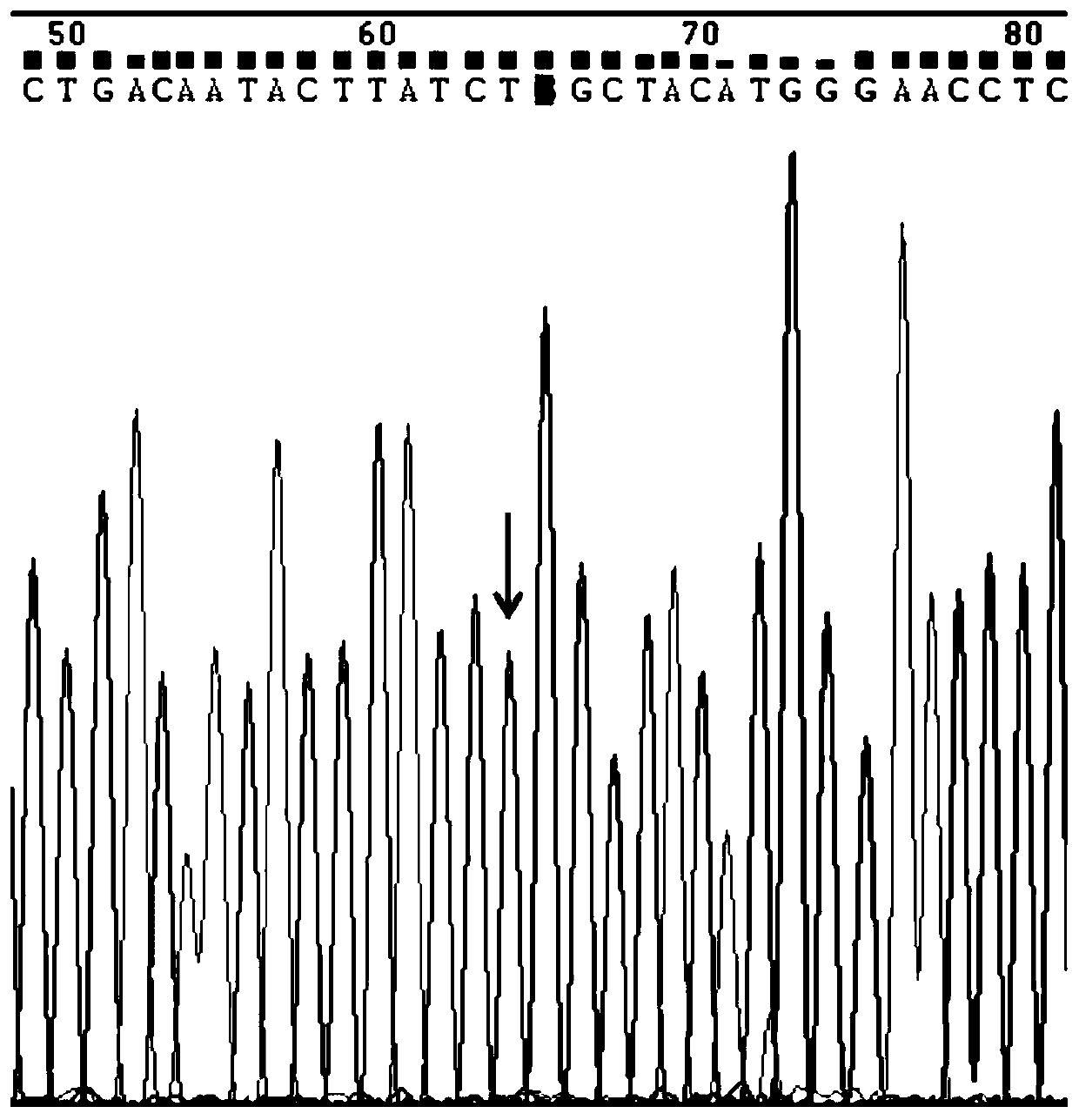
Image
Examples
Embodiment 1
[0037] 1. Extract the DNA from the sample:
[0038] Take 2ml of whole blood, and use the kit QIAamp DNA Mini Kit Cat.No.51304 to extract DNA. The specific steps are as follows:
[0039] (1) Add 200ul QIAGEN protease to a 15ml centrifuge tube, add 2ml blood sample, shake and mix;
[0040] (2) Add 2.4ml of AL buffer solution, shake and mix well, and incubate in a water bath at 70°C for 10 minutes;
[0041] (3) Add 2ml of absolute ethanol, mix upside down;
[0042] (4) Take half of the volume of the above liquid and add it to a 15ml QIAGEN separation column, centrifuge at 3000g for 3 minutes, and discard the filtrate;
[0043] (5) Repeat (5) for the remaining liquid in (3);
[0044](6) Add 2ml AW1 to the separation column, centrifuge at 3000g for 1 minute, and discard the filtrate;
[0045] (7) Add 2ml AW2 to the separation column, centrifuge at 3000g for 1 minute, and discard the filtrate;
[0046] (8) Centrifuge the separation column at 3000g for 1-2 minutes, and prepare a...
Embodiment 2
[0123] The preparation method of embodiment 2 prediction kit
[0124] (1) Using the above method to extract the genomic DNA from the peripheral blood of the subject to be tested.
[0125] (2) Use specific primers to amplify the fragments containing the three sites of rs13015714, rs9271588, and rs9277378. The following is the primer information as shown in Table 2:
[0126] Table 2: Amplification Primers
[0127]
[0128] (3) PCR reaction system and conditions:
[0129] The reaction system is shown in Table 3 below:
[0130] Table 3: Reaction system
[0131] Reagent Usage amount Final concentration 5xPCR buffer 2μl dNTP Mixture (2.5mM) 2μl 200uM magnesium chloride 1μl forward primer 6pmol 0.5uM reverse primer 6pmol 0.5uM Genomic DNA in the peripheral blood of the test subject 100-200ng DNA polymerase 1μl 1U / 20μl Deionized water Add to 20μl
[0132] Reaction Condition Table 4:
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com