Method for knocking out cytidine deaminase (cdd) gene in escherichia coli by utilizing CRISPR-Cas9 technology and application
A technology of cytidine deaminase and Escherichia coli, which is applied in the direction of microorganism-based methods, other methods of inserting foreign genetic materials, applications, etc., can solve adverse effects on human health and industrial production, affect efficient utilization of substrates, and separation operations Complicated problems, to achieve the effect of improving knockout efficiency, low cost, and high recognition efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Making Escherichia coli Competent Containing the Cas9 Plasmid Target Strain and Containing the cdd Gene
[0035] 1. The plasmid pCas9 was electrotransformed into Escherichia coli BL21(DE3) competent, and the target strain was screened out by kana resistance
[0036] Add 2-5 μg of plasmid pCas9 stored in the laboratory to 20-30 μL of Escherichia coli BL21(DE3) competent in the laboratory, pre-cool on ice for 2-5 minutes; add the pre-cooled electroporation cup, the conditions are: 200Ω, 2.0 kv electric shock; after electric shock, add 800-1000 μL LB medium immediately (the formula of LB medium is: peptone 10g / L, yeast powder 5g / L, sodium chloride 5g / L), 30℃, 2h culture; take 100μL medium The bacterial solution was coated with LB-resistant plate (kanamycin) and cultivated overnight at 30°C; single colony was picked, colony PCR was used to verify whether E. coli BL21(DE3) contained pCas9 plasmid, and the successfully constructed strain was named BL21(DE3)- cas9, ...
Embodiment 2
[0040] Example 2 Design and synthesis of mutant sgRNA
[0041] 1. Selection of N20 target site of cdd cytidine deaminase gene
[0042] The 399th position of the cdd PAM region of the cytidine deaminase gene was selected as the N20 target site by using the chop chop website, and the nucleotide sequence of the N20 target site is shown in SEQ ID NO.1.
[0043] 2. Design and synthesize mutant sgRNA
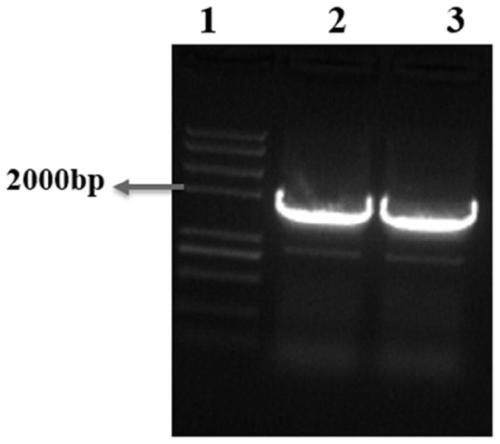
[0044] Use the snap gene software to open the sgRNA plasmid map purchased in the laboratory, replace the original N20 sequence with the selected N20 sequence, and design two primer sequences cdd-F1 (SEQ ID NO.2) and cdd-R1 (SEQ ID NO.3) Send it to Qingke Company for synthesis; use the sgRNA plasmid as a template and primers cdd-F1 and cdd-R1 for PCR amplification.
[0045] The PCR system is: primer cdd-F1 (10 μM) 2 μL, cdd-R1 (10 μM) 2 μL, 5×Buffer 10 μL, dNTPs (2.5mM) 5 μL, Fastpfu polymerase: 1 μL, sgRNA plasmid (100ng / μL) 2 μL, ddH 2 O to make up to 50 μL.
[0046] The reaction...
Embodiment 3
[0048] Example 3 Construction of cytidine deaminase gene cdd Donor DNA fragment
[0049] 1. Construction of upper and lower homology arms
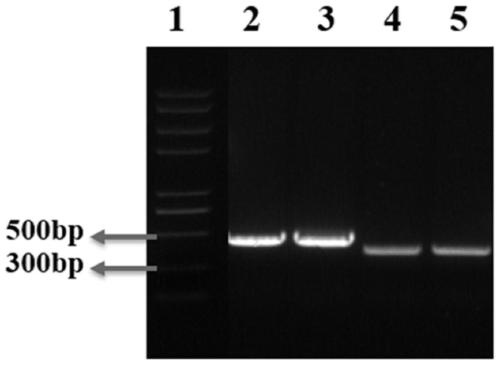
[0050] Use the NCBI website to find out the specific base sequence of the cytidine deaminase gene cdd, and import it using the snap gene software. Select the first 314bp of the gene as the upper homology arm, and the last 394bp as the lower homology arm, and design primers cdd-f-f (SEQ ID NO .5), cdd-f-r (SEQ ID NO .6), cdd-r-f (SEQ ID NO . NO.7), cdd-r-r (SEQ ID NO.8) were sent to Qingke Company for synthesis, using the Escherichia coli K12 series genome as a template, primers cdd-f-f and cdd-f-r, cdd-r-f and cdd-r-r were amplified by PCR respectively Add upper and lower homology arms M1, M2.
[0051] The PCR system is primer cdd-f-f / cdd-r-f (10μM) 2μL, cdd-f-r / cdd-r-r (10μM) 2μL, 5×Buffer 10μL, dNTPs (2.5mM) 5μL, Fastpfu polymerase: 1μL, Escherichia coli K12 series Genome (100ng / μL) 2 μL, ddH 2 O to make up to 50 μL.
[0052] The re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com