Method for analyzing human blood group genotype based on high-throughput sequencing, and application thereof
A technology of sequencing analysis and genotyping, applied in the field of bioinformatics, can solve problems such as incomplete serological testing, insufficient antibody reagents, difficulties in diagnosis and treatment of blood transfusion and immune diseases, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
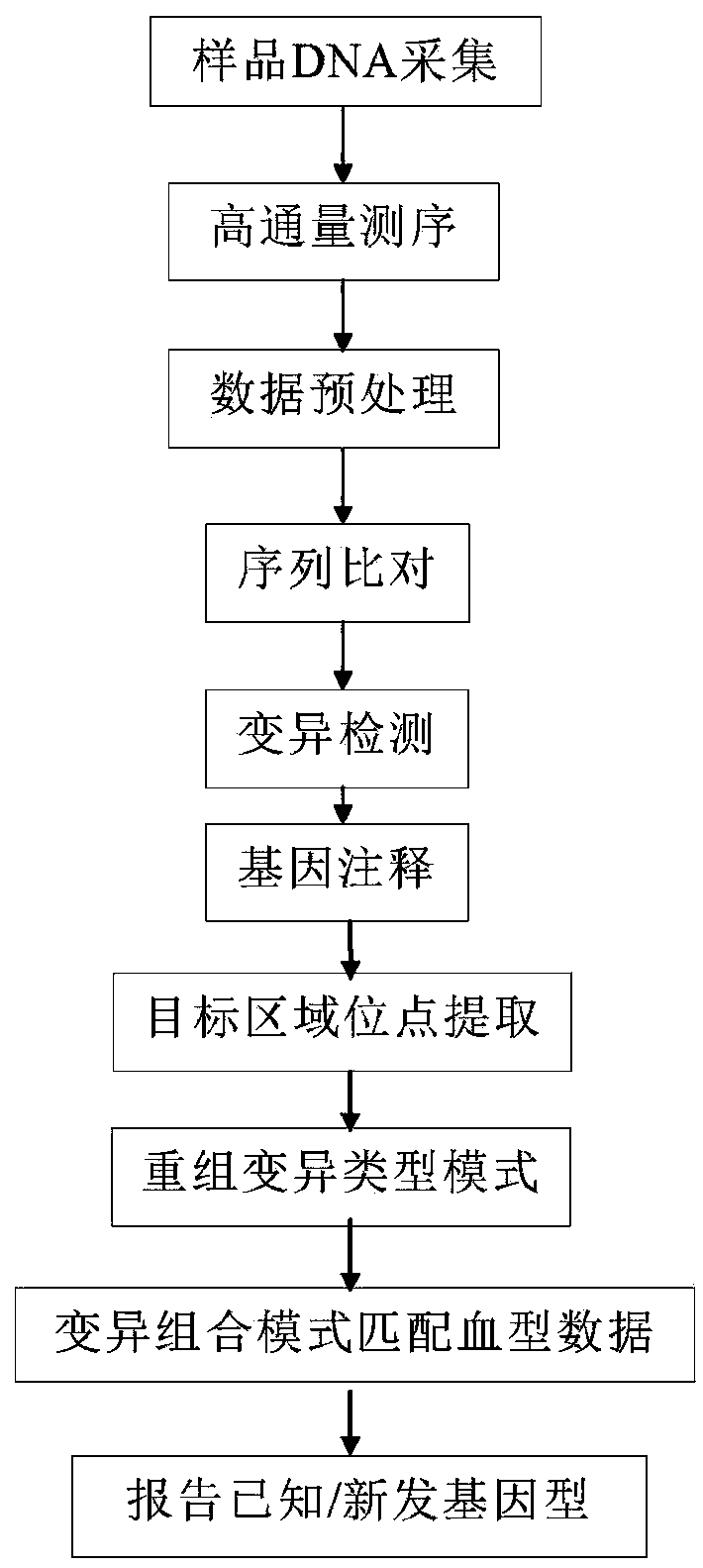
[0061] The technical route of this embodiment is as figure 1 shown. The specific implementation is as follows:
[0062] Randomly select 3,000 blood samples from voluntary blood donors and 175 representative samples and family investigation samples accumulated at ordinary times, and first use blood group serology to detect the type of MNS blood group antigen corresponding to red blood cell glycoprotein and the dose of red blood cell surface antigen (with Agglutination strength of antibody serum), select serologically representative samples to collect DNA for high-throughput whole genome sequencing.
[0063] Use high-throughput sequencing technology (NGS) to randomly fragment the genomic DNA of the selected sample, recover the DNA fragment of the required length (0.2-5kb) by electrophoresis, end repair - add adenylate deoxyribonucleotide, and connect the adapter , polymerase chain reaction, single-strand circularization, preparation of Nanoball-loading chip, using the method ...
s proportion 85
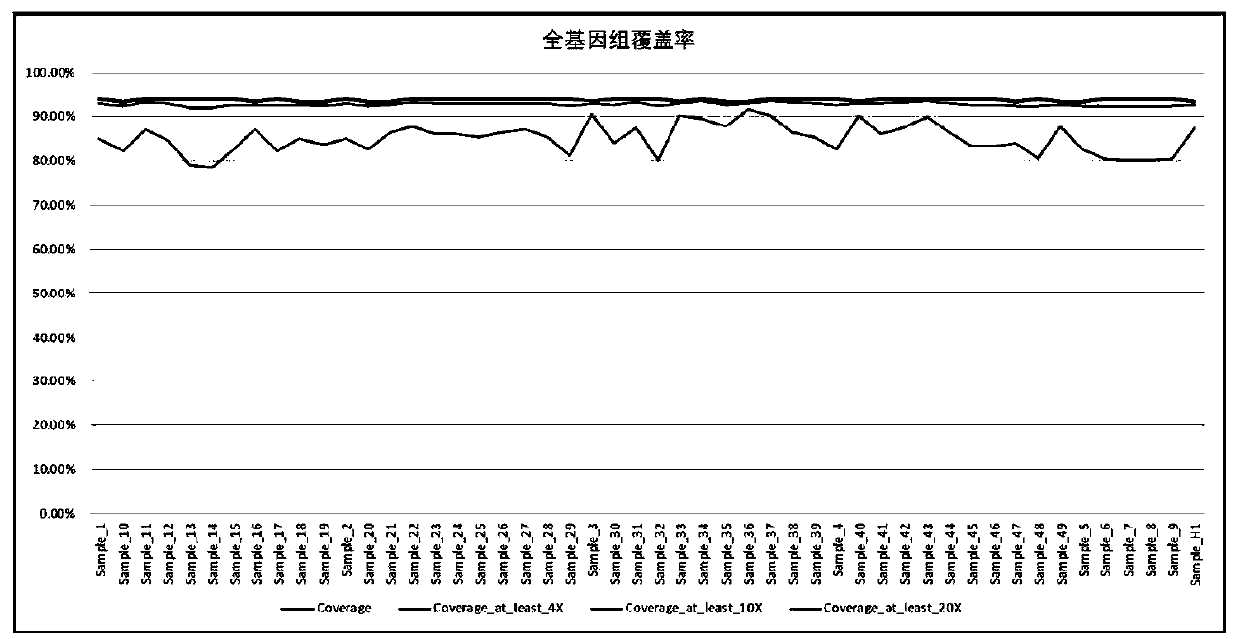
[0064] Obtain 30× whole genome sequencing (WGS) data, use SOAPnuke software to perform quality control analysis on off-machine data, filter low-quality Reads, obtain Q20 qualified Reads ratio of not less than 90%, and Q30 qualified Reads ratio of 85%. Focus on the analysis of GPA, GPB, GPE molecular corresponding GYPA, GYPB, GYPE gene and fusion gene GYP(A-B-A), GYP(B-A-B), GYP(A-B), GYP(A-B) The full-length sequence includes all information of exons, introns and UTR region sequences, such as image 3 shown. The inventors tested 30× whole genome sequencing data, the blood type gene detection rate was 100%, the average depth was 30.3×, and the average coverage rate was 100% for each gene. MNS blood group related genes GYPA / GYPB / GYPE The coverage rate is 98.3%, and the average depth is 25×, such as Figure 4 shown. It can meet the needs of research on the polymorphism of human blood group glycoprotein molecules, that is, the difficult blood group of MNS blood g...
Embodiment 2
[0069] Example 2 Cloud platform establishment
[0070] Such as Figure 9 As shown, the inventor has established a cloud platform for whole genome sequencing and blood typing, including an OSS cloud storage module, an ESC cloud service module and an RDS cloud data module.
[0071] The OSS cloud storage module unit, with a storage capacity of 10T, is used for sequencing data storage and can be used for long-term storage and reading and writing of data.
[0072] The ECS cloud data module is a cloud server equipped with 24-core 96GB running memory, 500GB high-efficiency SSD hard disk, and multiple genome-wide parallel computing capabilities to meet various biological information computing needs such as sequence comparison, mutation detection, and gene annotation.
[0073] The RDS cloud database will archive and build a database of the researched and newly released blood type genetic information, and establish a blood type information database.
[0074] The existing technology us...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com