Monomolecular mechanics method for measuring accurate length of human telomere
A technology of telomere length and molecular mechanics, which is applied in the field of measuring the precise length of human telomere using single-molecule force spectroscopy, can solve problems that have not been reported
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
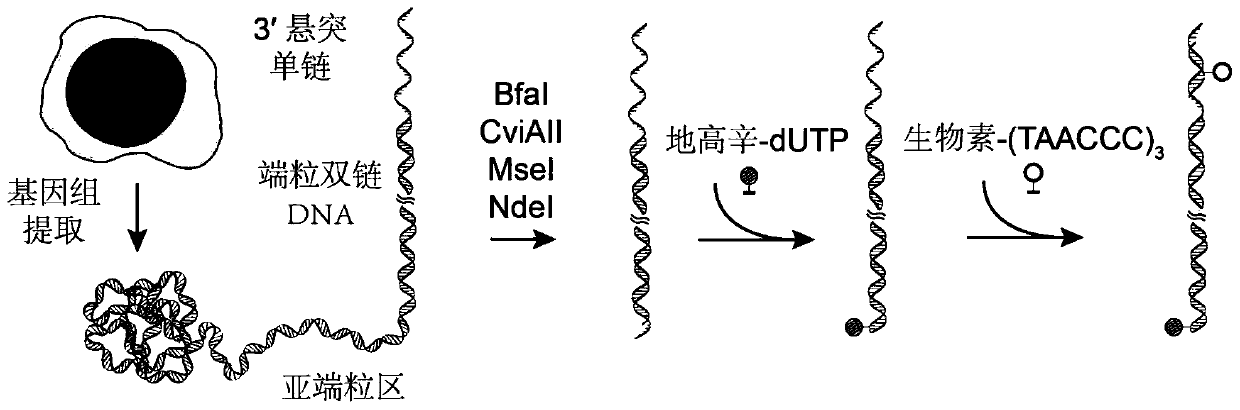
[0047] Example 1 Collection of human chronic myeloid leukemia cells K562 cells, extraction of cell genome
[0048] Collection of 1K562 cells
[0049] 1.1 Recovery and replacement of K562 cells
[0050] Take out 1 tube of cells (1.5ml, about 5×10 6 cells) were quickly thawed, and 2ml of complete medium was added (RPMI medium added with a final concentration of 10% fetal bovine serum, 100U / ml penicillin and 100mg / ml streptomycin solution). Centrifuge at 700 rpm for 5 minutes at room temperature, discard the supernatant, and add 5ml of complete medium to resuspend. Then the cells were transferred to 60mm Petri dishes and placed at 37°C, 5% CO 2 in the incubator. Cells attach and proliferate. After 24 hours of static culture, the medium was changed once. Take out the cell culture dish, place it in a biological safety cabinet, discard the medium supernatant, take 5ml of complete medium and slowly add it along the side wall of the culture dish. Continue culturing for 24 hours...
Embodiment 2
[0055] Example 2 Restriction endonuclease combinatorial digestion of genome, retention of telomeric DNA
[0056] The recovered genome was digested with restriction endonucleases (CviAII / NdeI / MseI / BfaI). Take 20 μg of genomic DNA in a 0.2ml PCR tube, add 5 μl of 10×Cutsmart buffer, 10 units of restriction endonuclease CviAII (product number: R0640L, NEB), make up to 50 μl with deionized water, and react at 25°C 12 hours. Add 10 units of restriction endonucleases NdeI (Cat. No.: R0111S, NEB), MseI (Cat. No.: R0525S, NEB) and BfaI (Cat. No.: R0568S, NEB) to the above reaction system, and react at 37°C for 12 hours . Heat at 80°C for 20 minutes to inactivate the above restriction endonucleases.
Embodiment 3
[0057] Example 3 Telomere DNA Double-Strand Region Affinity Modification and Single-Strand Region Affinity Probe Labeling
[0058] 3.1 Digoxigenin modification of telomeric DNA ends
[0059] Take 2 μg of genomic DNA digestion product, add 2 μl 10×0NEB buffer 2, and add dATP (product number: 4026Q, Baoriyi Biotechnology Co., Ltd.) and 0.2 mM Digoxigenin-11-dUTP (product number: 11093070910, Roche), and 10 units of Klenow Fragment (3′-5′exo-) polymerase (product number: M0212S, NEB), the volume was made up to 20 μl with deionized water. The reaction mixture was placed at 37°C for 12 hours to allow the sample to fully react.
[0060] 3.2 Biotin modification of telomeric DNA ends
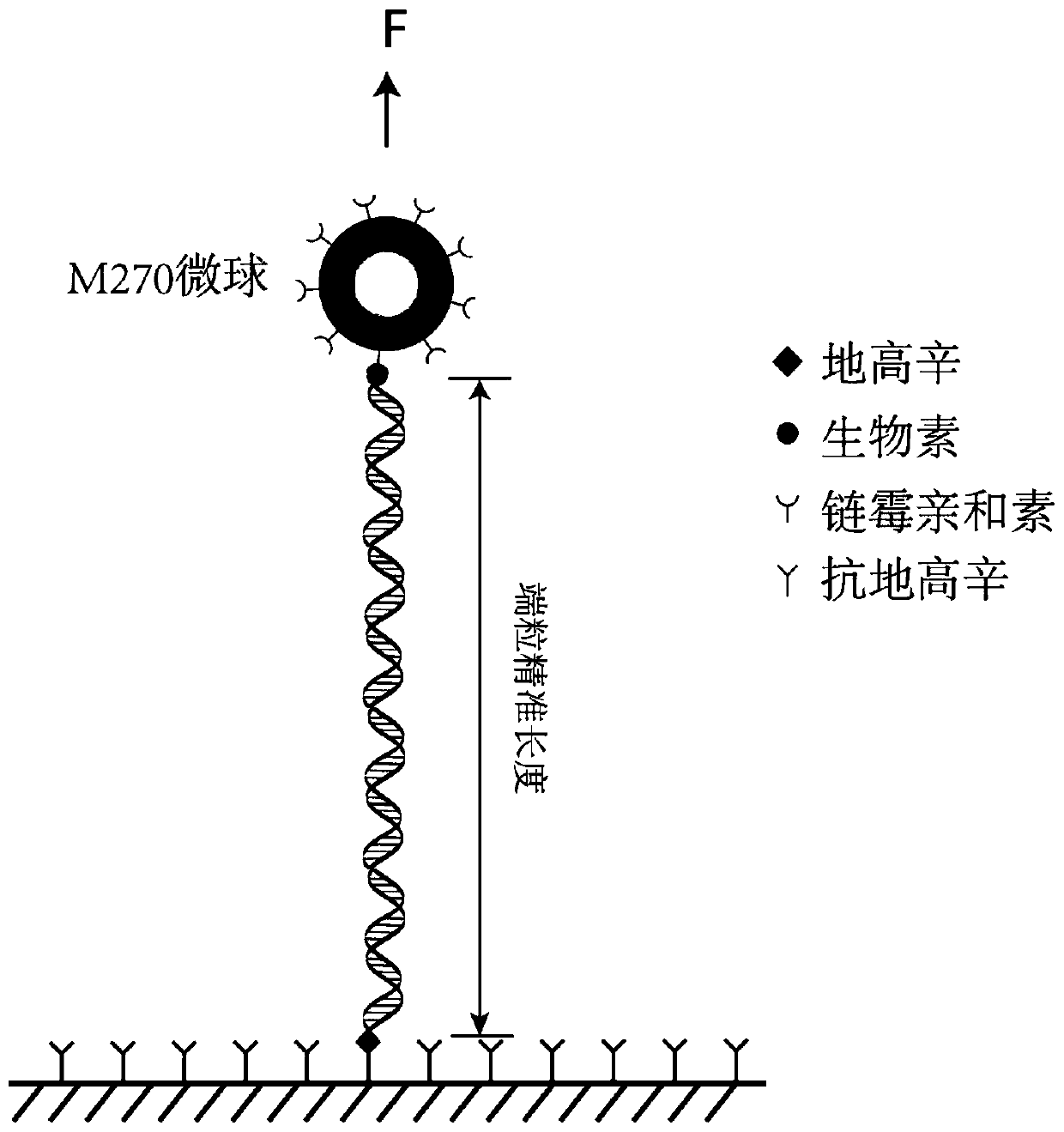
[0061] 10 μl (2.4 μg) of the product from the previous step was mixed with 2.4 μl (2.4 pmol) of a biotin-modified probe (Biotin-CCCTAACCCTAACCCTAA) and subjected to a gradient annealing test. The gradient annealing program was 75°C for 3 minutes; 75-25°C (decrease by 0.1°C every 8 seconds, a total of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com