Primer, method and reagent kit for detecting HBB gene mutation
A technology of kits and sequencing primers, which is applied in the fields of life science and biology, can solve the problems of tedious, time-consuming and labor-intensive detection of gene HBB, and achieve the effects of simplifying operation steps, saving detection costs, and reducing detection costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Kits for detecting gene HBB mutation sites, including: tissue DNA extraction kit (for example, using Tiangen Bio’s DNA extraction kit); absolute ethanol; amplification system PCR reaction solution, sequencing system reaction solution, positive control , negative control substance and blank control substance, wherein
[0057] Amplification system PCR reaction solution includes: 2×PCR Buffer; 2mMdNTPs; KOD FX DNA Polymerase (1U / μl); at least one pair of amplification primers are used to amplify the gene HBB, and the amplification primers are selected from HBB-1_2F / HBB-1_2R , HBB-3_6F / HBB-3_6R, HBB-7_9F / HBB-7_9R, HBB-10F / HBB-10R, the base sequence of which is:
[0058] HBB-1_2F: TGATGTGGAACCAGAAAGCTGTAAAACGACGGCCAGT
[0059] HBB-1_2R: GGGCAGCATAGCAAGAACAACAGCTATGACCATG;
[0060] HBB-3_6F: TCCCACCTCAGCCTCAAGTTGTAAAACGACGGCCAGT
[0061] HBB-3_6R: ACCCGCCTCATAGCAATGAACAGCTATGACCATG;
[0062] HBB-7_9F: ACAGCCAGTGATGCTACCGTGTAAAACGACGGCCAGT
[0063] HBB-7_9R:ACCAGCCTCCACCA...
Embodiment 2
[0075] Example 2 Blood sample DNA detection process
[0076] (1) Genomic DNA extraction from blood:
[0077] 1) Draw 500uL of blood and add 1000uL of red blood cell lysate, mix it upside down, place it at room temperature for 5 minutes, and then mix it upside down several times during this period, then centrifuge at 3000rpm for 5min, absorb the supernatant, leave the white blood cell precipitate, add 200uL buffer GA, shake until thoroughly mixed;
[0078] 2) Add 20 μl proteinase K solution and mix well;
[0079] 3) Add 200 μl buffer GB, mix thoroughly by inversion, place at 70°C for 10 minutes, the solution should become clear, and briefly centrifuge to remove water droplets on the inner wall of the tube cap;
[0080] 4) Add 200 μl of absolute ethanol, shake and mix well for 15 seconds. At this time, flocculent sediment may appear, and briefly centrifuge to remove water droplets on the inner wall of the tube cap;
[0081] 5) Add the solution and flocculent precipitate obtai...
Embodiment 3
[0114] Three clinical whole blood samples were taken, and the whole exon mutation of the HBB gene related to β-thalassemia was detected in each sample. The genome was extracted, reagents were prepared and tested according to the method described in Example 2. For each sample, 2 μl of the extracted genomic DNA was added to the PCR reaction solution of the amplification system, and positive, negative, and blank control experiments were performed at the same time. A 96-well ordinary PCR machine can detect 46 samples at the same time, each sample has 2 repetitions, a positive control, a negative control and a blank control. The detection time is 160 minutes.
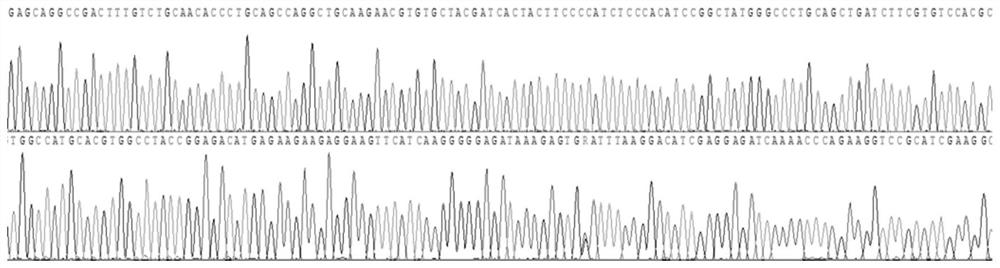
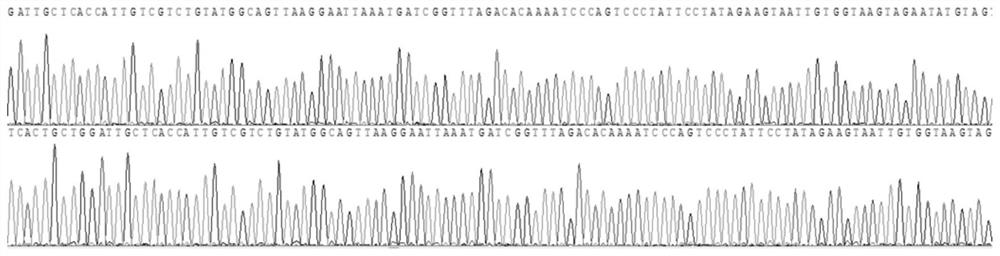
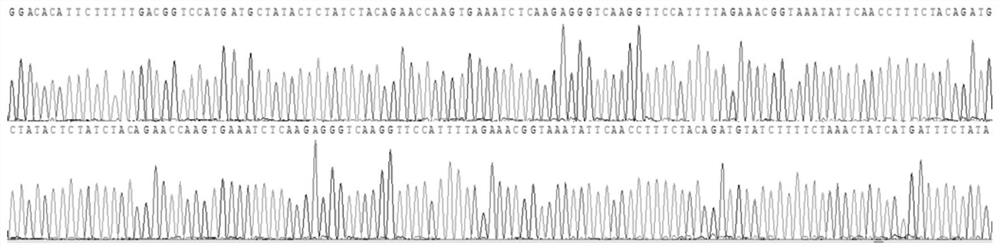
[0115] In addition, the sequencing results of sample 1 are as follows Figure 1-4 All shown are wild type. The sequencing results of samples 2 and 3 were also wild type.
[0116] It can be seen from the detection results that the primers of the present invention have included all the exons of the HBB gene to be detected,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com