Specific reversible newborn protein enrichment method
A protein and species-specific technology, applied in the field of protein enrichment, can solve the problems of reducing the ionization efficiency of peptides, interference of non-specifically adsorbed proteins, and affecting protein identification, etc., and achieves the effects of high efficiency, mass spectrometry compatibility, and convenient and reliable separation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
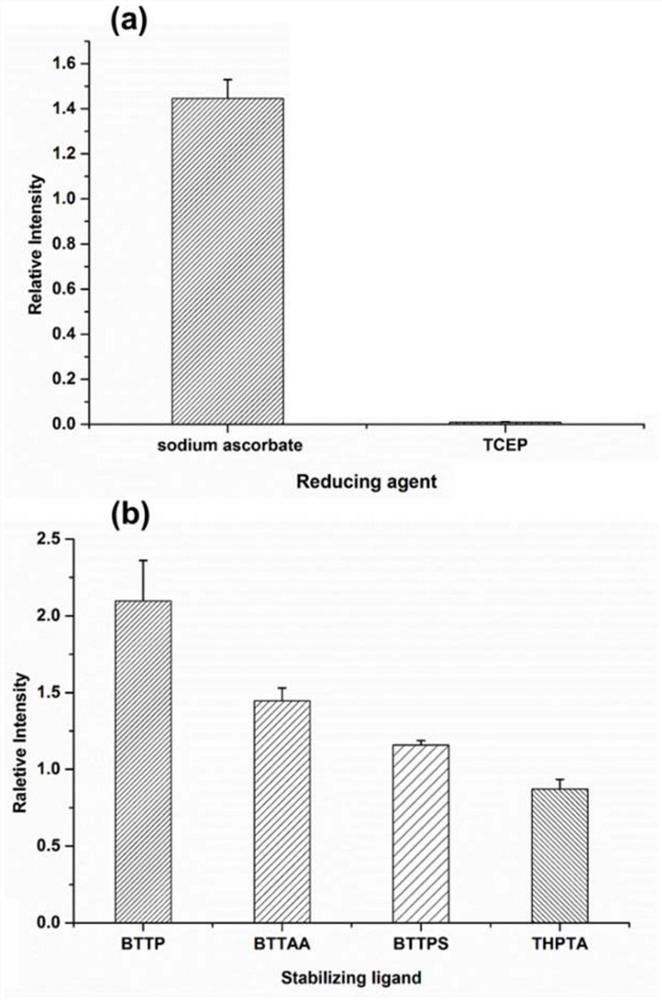
Method used
Image
Examples
Embodiment 1
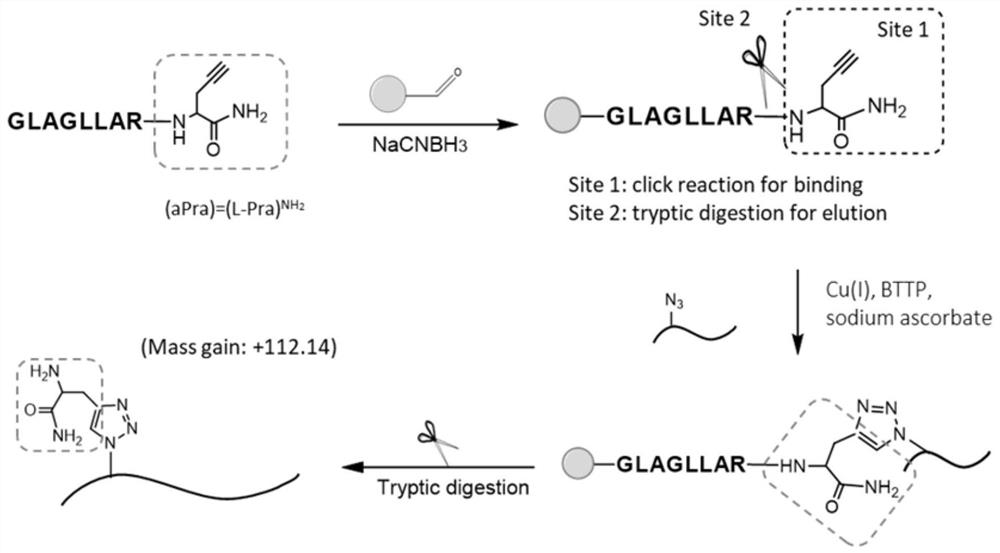
[0080] Example 1: The method established based on the cleavable bioorthogonal reversible tag (CBOT) enriches the synthesized azido peptides. The enrichment principle of the CBOT tag is shown in figure 1 , the specific process of the experiment is as follows:
[0081] (1) Take 10 μL of commercial aldehyde resin, add 10 μg of synthetic alkynyl peptide GLAGLLAR (aPra), and add reducing agent NaCNBH3 to PBS with pH 7.4 to cause reductive alkylation reaction to obtain alkynyl functionalization Resin materials, referred to as alkyne-based resins;
[0082] (2) Take the synthesized alkyne-based resin, add 10 μg of azide peptide (m / z 860), and CuAAC catalyzes the covalent binding to the alkyne-based resin;
[0083] (3) Trypsin hydrolysis, the released target peptide was added with aPra group, detected by MALDI, the molecular weight increased to 112.12 (m / z 972).
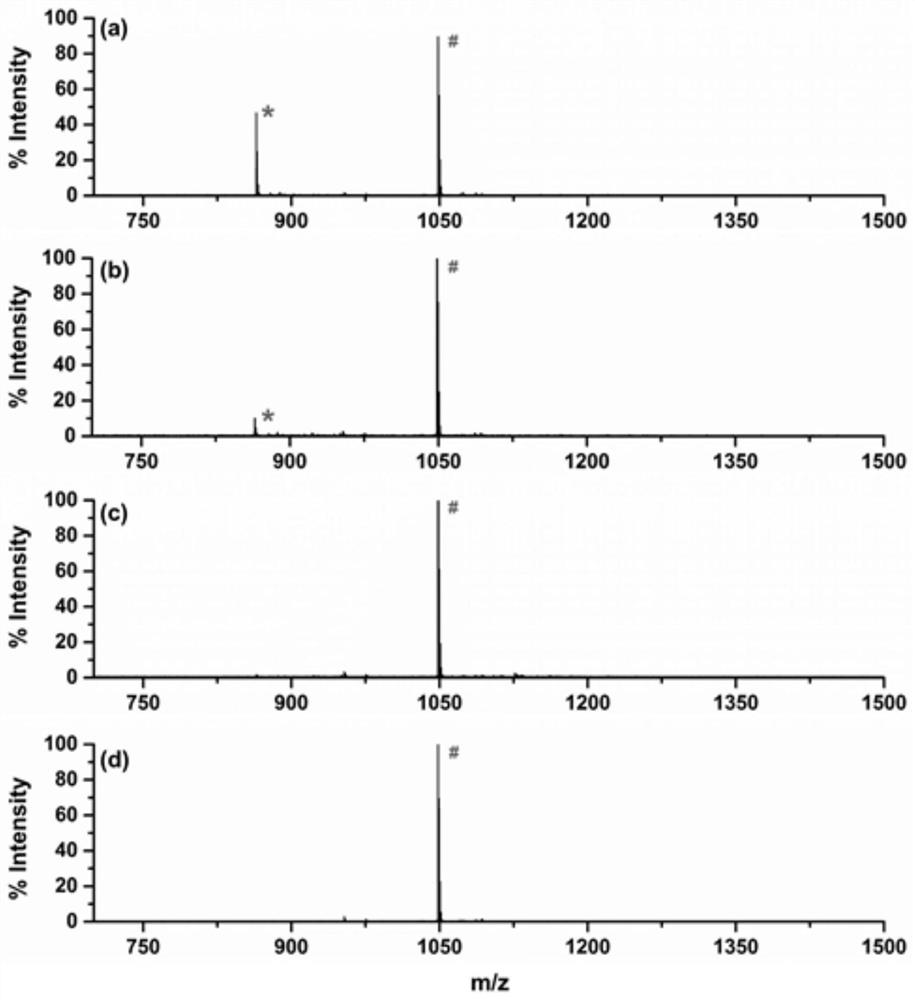
[0084] figure 2MALDI spectrum for evaluating the capacity of alkyne groups on functionalized resin materials. After di...
Embodiment 2
[0087] Example 2: The cleavable label based on bioorthogonal reaction combined with trypsin digestion in Example 1 is used for the enrichment and identification of nascent proteins in 293T cells. The established enrichment and identification process for nascent proteins is shown in Figure 5 , the specific experimental procedure is as follows:
[0088] (1) AHA-labeled 293T cells, extract the whole sample: 1mM AHA, culture for 4 hours, collect cells to lyse and extract protein;
[0089] (2) Prepare 10 μL of alkyne-based resin according to step (1) in Example 1;
[0090] (3) According to step (2) in Example 1, 10 μg of azide peptides were replaced with 500 μg of enzymatic peptides of AHA-labeled whole protein to complete the CuAAC catalytic connection;
[0091] (4) Strong elution conditions to remove non-specifically adsorbed proteins. Wash sequentially with 200 times volume of elution reagents, respectively 1% SDS, 8M urea, PBS and 50% acetonitrile solution.
[0092] (5) Try...
Embodiment 3
[0096] Example 3: The process of CBOT enrichment of nascent proteins in Example 2 combined with light and heavy isotopes AHA / hAHA was used for quantitative research on nascent proteins in HEK293T cells. The specific experimental process is as follows:
[0097] (1) 293T cells were labeled with AHA and hAHA, incubated for 4 hours, and the cells were collected and lysed;
[0098] (2) Separately extract and quantify the protein, and mix the two samples with equal mass at 1:1;
[0099] (3) according to the flow process in protein embodiment 2 enrichment nascent protein;
[0100] (4) Added variable modifications: AHA(Met) and hAHA(Met) in the mass spectrometry library settings, and set mass changes +107.07 and +113.17 as variable modifications for protein identification and assignment. At the same time, the protein was quantitatively analyzed according to the AHA peptide and the corresponding hAHA peptide.
[0101] Figure 8 It is the omics result of using the CBOT method in Exa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com