Method for efficiently and specifically removing plasmids in bacteria by utilizing integrated suicide vector
A suicide vector, specific technology, applied in the field of gene editing, can solve the problem that the efficiency of plasmid elimination cannot be guaranteed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
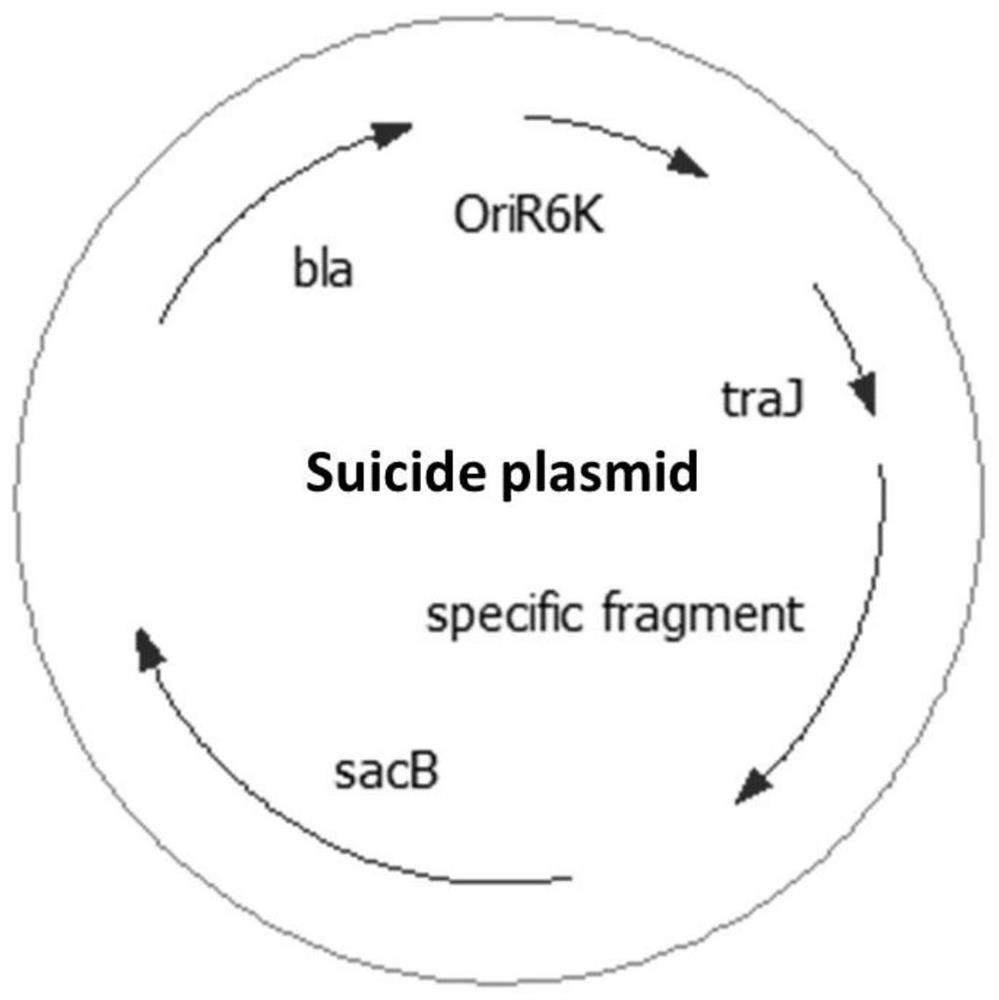
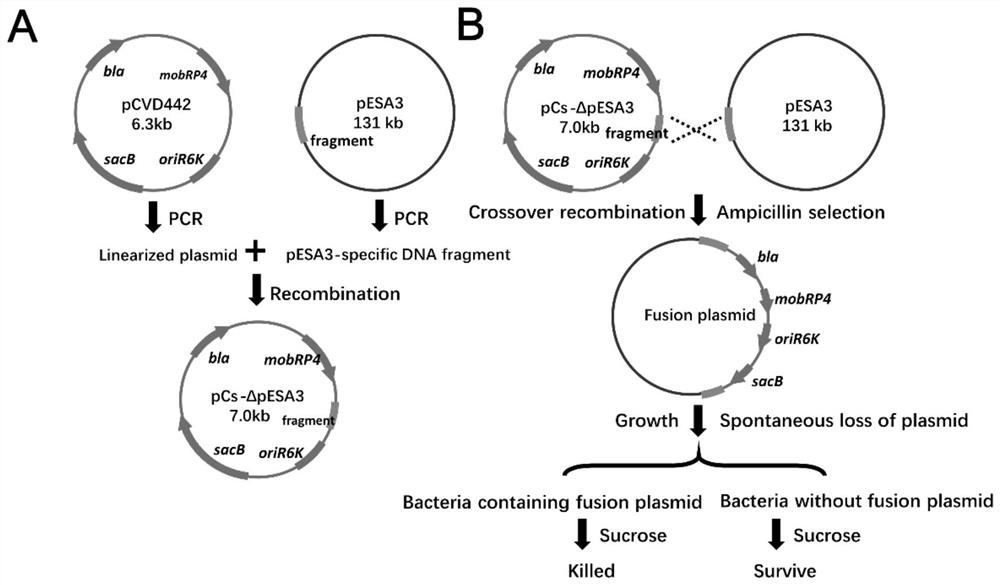
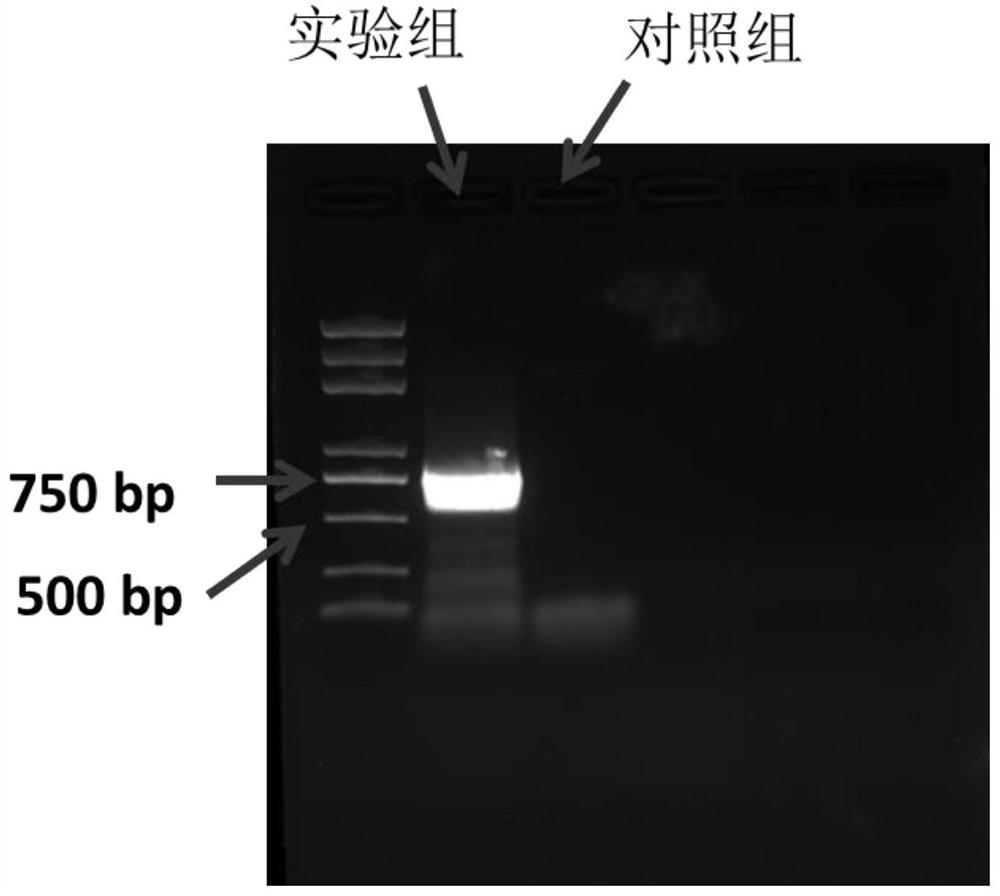
[0038] A method for efficient and specific removal of plasmids in bacteria using an integrated suicide vector, the vector pCs-ΔpESA3 displays a pESA3-specific fragment and the ampicillin resistance gene bla, replication initiator oriR6K, sucrose-sensitive marker sacB and broad host range mobility Region localization of mobRP4. pCVD442 was linearized by PCR and ligated with pESA3-specific DNA fragment to obtain plasmid pCS-ΔpESA3.
[0039] The plasmid pCS-ΔpESA3 was extracted from S17-1λpir Escherichia coli and transformed into Enterobacter sakazakii BAA-894. Through genetic recombination of the pCS-ΔpESA3 plasmid and the integration of the endogenous plasmid pESA3 of BAA-894, the Enterobacter sakazakii BAA-894 containing the integrated plasmid was obtained through ampicillin selection, and cultured in a non-resistant medium caused the spontaneous loss of the integrated plasmid of some bacteria , sucrose exposure selectively kills bacteria harboring integrated plasmids for the...
Embodiment 2
[0061] In order to verify the generality of the method, another integrated suicide vector was constructed in an attempt to knock out the plasmids leading to multi-drug resistance in a variety of bacteria.
[0062] Cronobacter sakazakii GZcsf-1, Shigellaflexneri strain M2901, Salmonella enterica subsp.enterica serovarTyphi strain, Citrobacter freundii strain Iona 4 and pneumonia Klebsiella pneumoniae strain555 all have antibiotic resistance, and their drug resistance is considered to be related to the plasmids they carry, which are pGW1 plasmid (340kbp) of Enterobacter sakazakii, pB4878 plasmid (55kbp) of Salmonella, pM2901 plasmid (81 kbp) for Shigella flexneri, pCFI-2 plasmid (24 kbp) for Citrobacter flexneri and pSCKLB555-1 plasmid (247 kbp) for Klebsiella pneumoniae. There are homologous segments in these 5 plasmids, and these 5 plasmids may have a common origin. Utilizing the homologous segments in these five plasmids, the homologous segments were cloned into the suicide ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com