Primer group for simultaneously identifying wild strain and gene deletion strain of African swine fever based on multiple qPCR technology and test kit
An African swine fever and gene deletion technology, applied in recombinant DNA technology, microbial assay/test, biochemical equipment and methods, etc., can solve the problem of inability to distinguish between ASF wild virus strains and gene deletion vaccine strains, and achieve low cost , the effect of saving costs and reducing pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] 1. Design of specific primers and specific probes: Download multiple ASFV full genes (HLJ / 2018, SY18, N10 / 2018, AnhuiXCGQ) in Genbank, and use the BioEdit software package to perform Clustalw comparison. According to the p72 gene conservation of the above viruses Sequence, using Primer 5.0 software to design specific TaqMan probes and primers. According to the deletion sequence of the attenuated live vaccine strain HLJ / 18-7GD of the Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (HLJ / 2018 strain deletion 73394-74476 nucleotides (CD2v) and 27942-35500 nucleotides (MGF) ) and the ASFV gene deletion vaccine strain (SY18 strain lacks 73368-74452 nucleotides (CD2v) and 27942-35500 nuclear (MGF)) consensus deletion fragment, using Primer 5.0 software to design specific TaqMan probes and primers for CD2v gene and MGF gene.
[0060] 2. Synthesis of primers and probes: synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
[0061] 3. See...
Embodiment 2
[0072] Multiplex real-time fluorescence method validation
[0073] 1. Specificity Verification
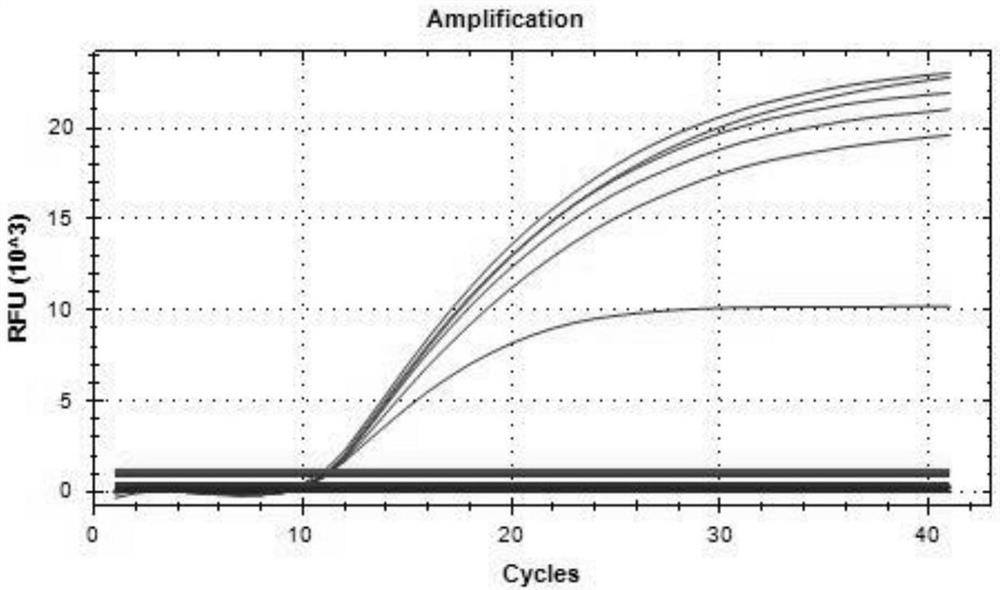
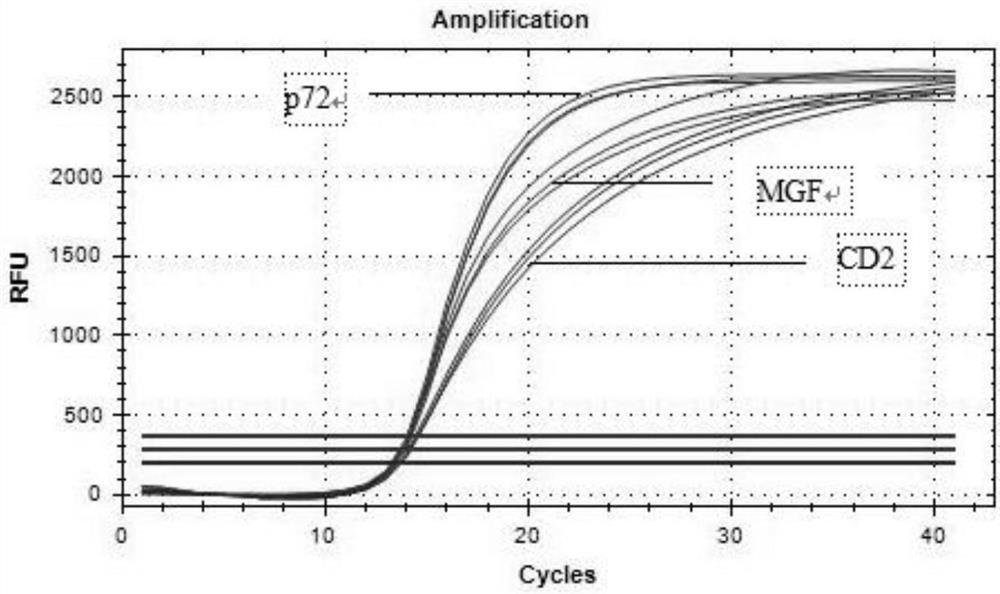
[0074] Utilize kit method of the present invention to respectively with the DNA of ASFV wild strain DNA and gene deletion vaccine strain DNA, porcine parvovirus (PPV), porcine pseudorabies virus (PRV) and porcine circovirus type 2 virus (PCV2) and pig breeding The specificity of primers and probes was verified by multiplex real-time fluorescent PCR amplification with the cDNA of respiratory syndrome virus (PRRSV) and porcine epidemic diarrhea virus (PEDV) as templates. The results are shown in Table 3, and the results show that the designed primers and probes of the present invention have strong specificity.
[0075] Table 3 specificity verification test
[0076]
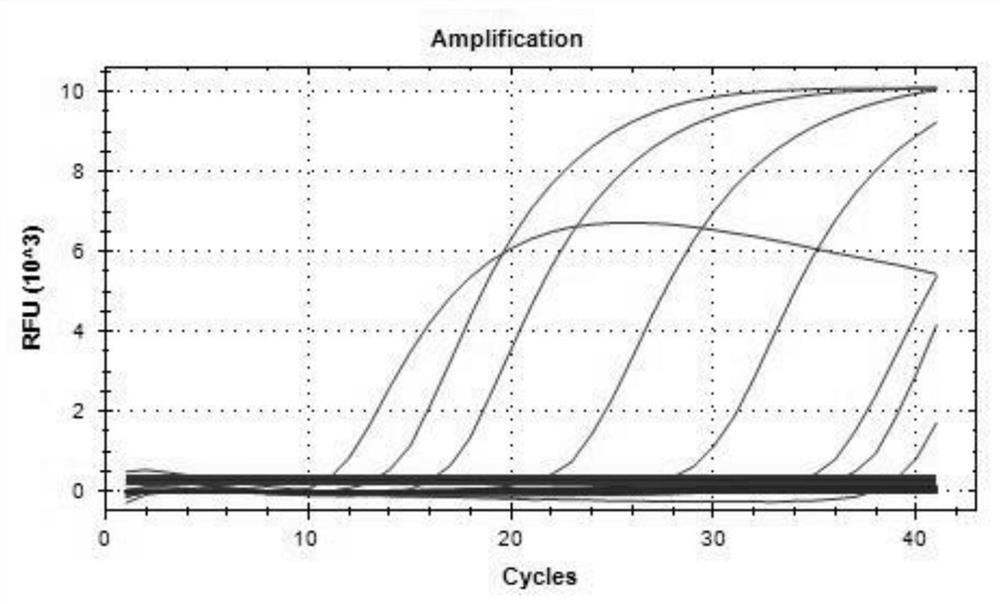
[0077] 2. Sensitivity evaluation
[0078] Quantify the positive standard products of ASFV p72 gene, CD2v gene and MGF gene to 107 copies / μL respectively, and serially dilute them by 10 times to 1.0×10 6 , 1.0×10 ...
Embodiment 3
[0081] Clinical suspicious sample detection
[0082] The multiple real-time fluorescent PCR detection method established by the present invention to simultaneously identify wild African swine fever strains and gene deletion vaccine strains detects 12 clinically suspicious samples, and the sample types include pig lungs, lymph nodes, tissues and serum. At the same time, positive samples were subjected to ordinary PCR detection and parallel sequencing analysis.
[0083] The results are shown in Table 4, and the results show that the method established by the present invention is consistent with the ordinary PCR detection results and sequencing results, and the method is accurate and reliable.
[0084] Table 4 Test results of clinical samples
[0085]
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com