Gastric mucosa lesion protein molecular typing, lesion progression and gastric cancer related protein marker and method for predicting lesion progression risk
A technology of molecular typing and gastric mucosa, applied in the field of clinical tumor medicine, can solve the problems of lack of multiple comparison correction verification of differential proteins, proteomic changes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0106] Example 1. Obtaining protein expression profile data of gastric mucosal tissue samples obtained from clinical gastroscopy biopsy
[0107] The experimental samples were 169 gastroscopic biopsies of gastric mucosa tissue samples from the high-incidence site of gastric cancer in Linqu, Shandong Province and the Fifth Medical Center of the PLA General Hospital.
[0108] Protein extraction and analysis were performed on 169 gastric mucosal tissue samples from clinical gastroscopy biopsy. Through this step, the proteome data set corresponding to each sample was obtained, including the type, quantity and quantitative value of each protein.
[0109] 1. Lysis solution formula:
[0110] 1% (w / v) DOC (Deoxycholic acid), 10mM TCEP,
[0111] 40mM 2-chloroacetamide (CAA), 100mM Tris, pH 8.5.
[0112] 2. Operation steps
[0113] 1. Material collection: gastroscope samples, which are stored in clean EP tubes after collection;
[0114] 2. Cleavage sample: add 500uL lysate and homoge...
Embodiment 2
[0135] Part I: Proteomic Molecular Typing
[0136] Proteomic molecular typing of gastric mucosal lesions based on the data in Example 1, the specific steps are as follows:
[0137] 1) Protein expression profile preprocessing and experimental filtering
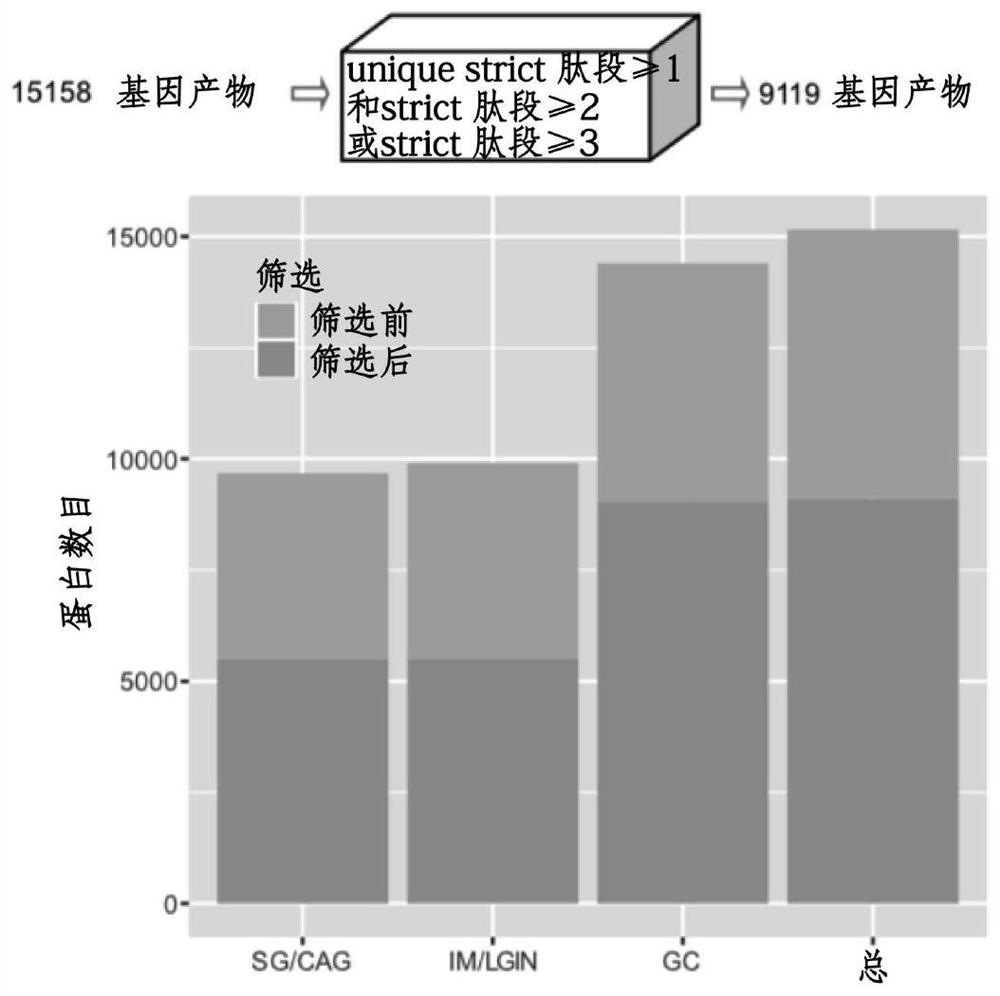
[0138] a) High-confidence protein screening: Quantitative proteins are required to contain at least one unique peptide with a Mascot ion score greater than or equal to 20, and at least two peptides with an ion score greater than or equal to 20, or three ions Peptides with a score greater than or equal to 20;
[0139] b) Quantitative data standardization based on the sum: using the non-labeled quantitative iBAQ method based on the peak area, the iBAQ value of a protein is the sum of the peak areas of all corresponding peptides of the protein / theoretical peptide number, each identified by calculation The ratio of the protein iBAQ value to the sum of all identified protein iBAQ values was used to normalize the data to obtain a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com