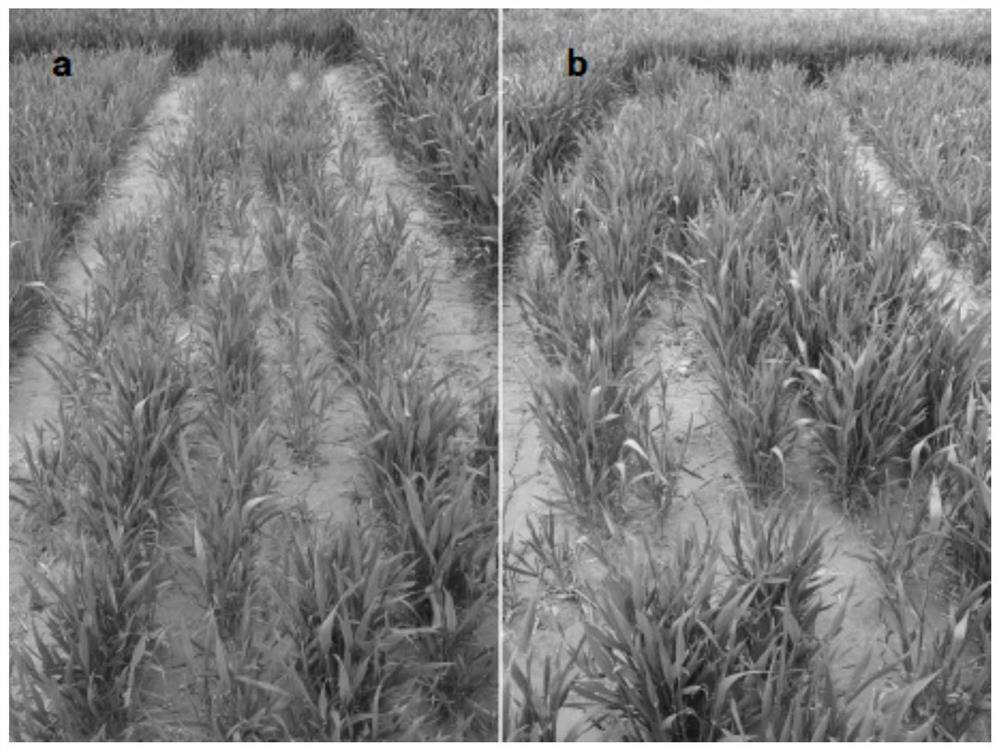
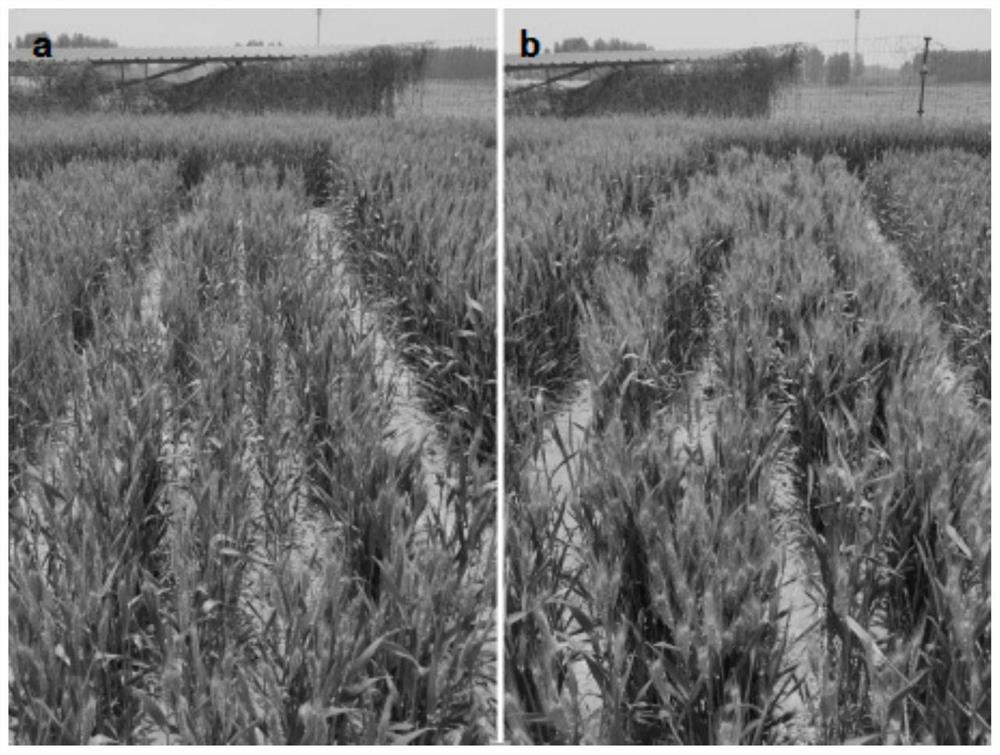
Chaetomium globosum and application thereof in promoting growth and increasing yield of wheat
A technology of Chaetomium globosa and wheat grains, applied in the direction of application, microbial-based methods, and microbial-using methods, can solve problems that have not been reported, and achieve the effect of increasing yield and increasing chlorophyll content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Isolation of Chaetomium globosa Strain 12XP1-2-3
[0023] At the wheat grain filling stage, the uninfected plants in the wheat fields were collected in Xiping County, Henan Province and put into paper bags. The samples were stored in a refrigerator at 4°C. Rinse the root of the sample, and then take 2-3cm of wheat root tissue, disinfect the surface with 75% absolute ethanol for 10-30s, disinfect with 1% NaCIO for 1.5min, rinse with sterile water for 3 times, and blot dry with sterile filter paper After hydration, cut into small sections of about 0.5 cm, and place them on a potato dextrose agar medium (PDA, raw material composition: 300 g of peeled potatoes, 20 g of glucose, 20 g of agar and 1000 ml of distilled water) with sterile tweezers. After 5-7 days of cultivation under the same conditions, pick off the edge mycelia and grow on a new PDA plate. After 5-7 days under the same conditions, pick off the mycelium pieces with uniform colonies and place them on a PDA sla...
Embodiment 2
[0025] Extraction of DNA from Chaetomium globosa Strain 12XP1-2-3
[0026] Pick the chaetomium strain 12XP1-2-3 onto the PDA medium, and after culturing at 20°C for 5-7 days, pick 5 edge mycelium pieces and place them evenly on the PDA plate covered with sterilized cellophane. After 3-5 days, scrape the mycelium with a sterilized small shovel, freeze it with liquid nitrogen, and store it in a refrigerator at -20°C. When extracting DNA, take out 20-25 mg mycelium and put it into a pre-cooled 1.5ml EP tube; add a little liquid nitrogen and grind the mycelium to powder in the EP tube with a pre-cooled iron nail; add 500 μl Extraction Buffer (50mM Tris -Cl (pH 8.0), 150mM NaCl, 100mM EDTA (8.0)), shake the suspended pellet on a vortex shaker and mix well; , mixed upside down, placed in a water bath at 37°C for 1-3 hours; then added 75 μl of 5M NaCl, mixed upside down; added 65 μl of CTAB / NaCl (10% CTAB, 0.7M NaCl) solution, bathed in water at 65°C for 30 minutes ;Add an equal vo...
Embodiment 3
[0028] Molecular identification of Chaetomium globosa strain 12XP1-2-3
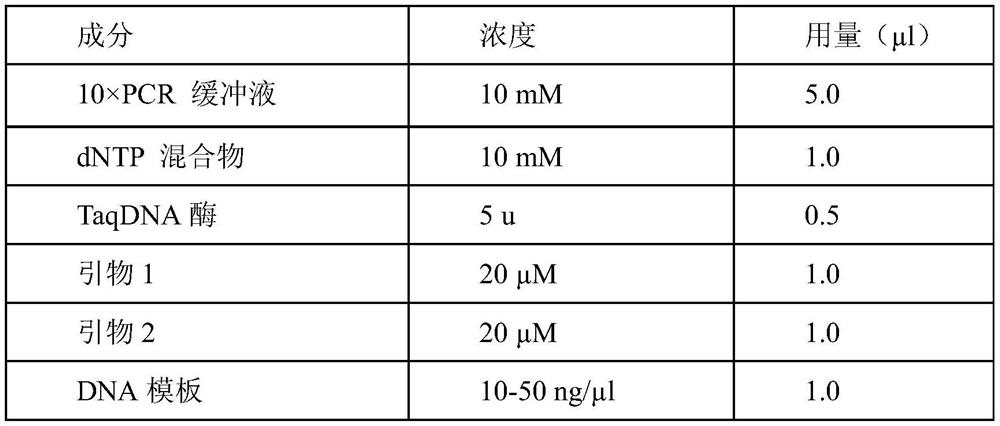
[0029] In this test, primers ITS1 (5'-TCC GTA GGT GAA CCT GCG G-3') and ITS4 (5'-TCC TCCGCT TAT TGA TAT GC-3') were used to amplify the strains containing ITS1, 5.8S rDNA and ITS2 of DNA fragments. The system refers to Daval et al. (2010). Amplification reaction conditions: The PCR reaction program is: 95°C for 3min; 95°C for 45s, 50°C for 30s, 72°C for 1min, 35 cycles; 72°C for 10min. After the PCR products were sequenced, BLAST sequence alignment analysis was carried out in NCBI. The result of the sequence alignment shows that the biocontrol bacterium 12XP1-2-3 is a globular hair shell.
[0030] Table 1. PCR reaction system
[0031]
[0032]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com