Primer group for amplifying B lymphocyte immune repertoire and application of primer group
A technology of B lymphocytes and immune repertoire, applied in recombinant DNA technology, microbial assay/test, biochemical equipment and methods, etc., can solve the problem of unclear pathogenesis of IgAN, achieve high coverage and reduce secondary Structure generation, specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] This embodiment provides a method for expanding the information of the immune repertoire of B lymphocytes in IgAN patients.
[0057] 1. Patient recruitment and screening
[0058] In this example, the whole blood samples and kidney biopsy samples of 13 IgAN patients were collected, aged between 22 and 62 years old (the median was 26), and 7 normal people were recruited at the same time. According to the method recommended by WHO, blood and urinalysis and renal biopsy were performed on the main components. This study was conducted in accordance with the guidelines of the Endocrinology Department of Shenzhen Nanshan Hospital and was approved by the local medical ethics committee. All patients provided written informed consent.
[0059] 2. PBMC separation
[0060] Dilute 10 mL of blood with an equal amount of PBS and carefully load it into 5 mL of lymph preparation solution. The ratio of sample volume to lymph preparation medium is 2:1. All reagents used are purchased fr...
Embodiment 2
[0082] In this example, data analysis is performed on the BCR library prepared in Example 1.
[0083] A total of 18,976,912 pairs of end reads were generated by the Illumina sequencer, and overlapping paired-end reads were merged using FLASH software to obtain 16,376,727 original reads;
[0084] Merged sequence reads were aligned to V, D, and J gene germline references using IgBLAST, and constant regions were mapped to IgA, IgD, IgE, IgG, and IgM, respectively;
[0085] The reference sequences of the V, D and J gene germlines were obtained from IMGT, and reads with low homology (<70%) to the germline reference line were excluded. After filtering, 11,114,426 reads were obtained for further analysis;
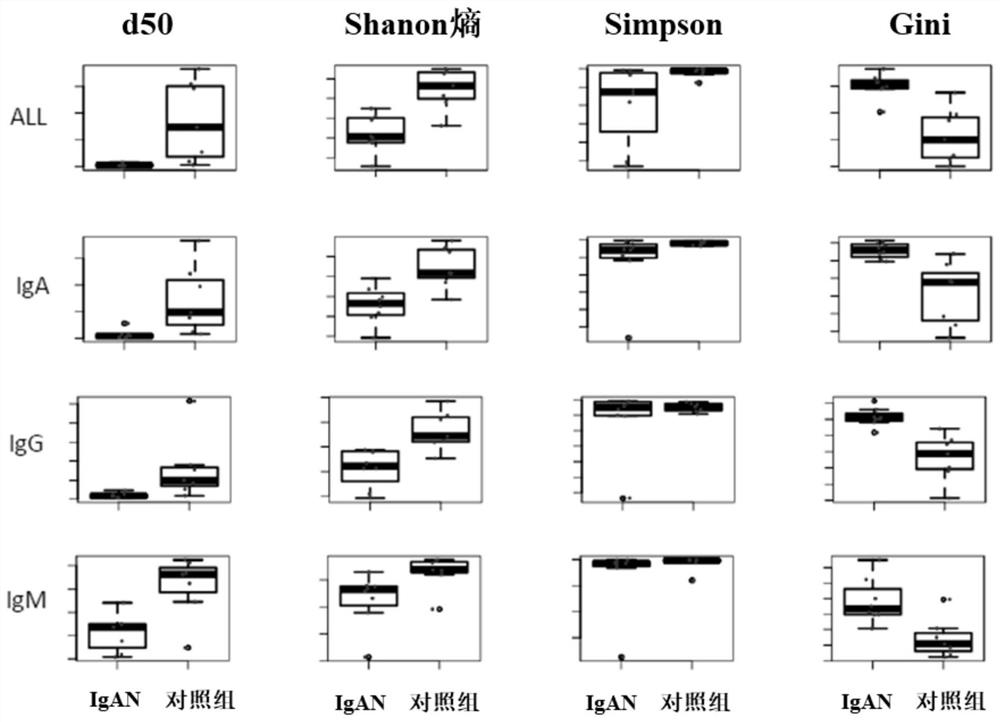
[0086] According to the definition of IMGT, the start and end position, reading frame and productivity of the CDR3 region were determined. The overexpression level of IgAN samples was analyzed by software MixCR. Diversity of the antibody repertoire was evaluated with normalized ...
Embodiment 3
[0088] The length distribution of CDR3 in IgAN and normal candidate (NC) populations was analyzed in this example. The IgAN and NC libraries were constructed according to the method described in Example 1, and the data analysis method was analyzed according to the method described in Example 2.
[0089] (1) Analysis of the length distribution of CDR3 in IgAN and NC populations
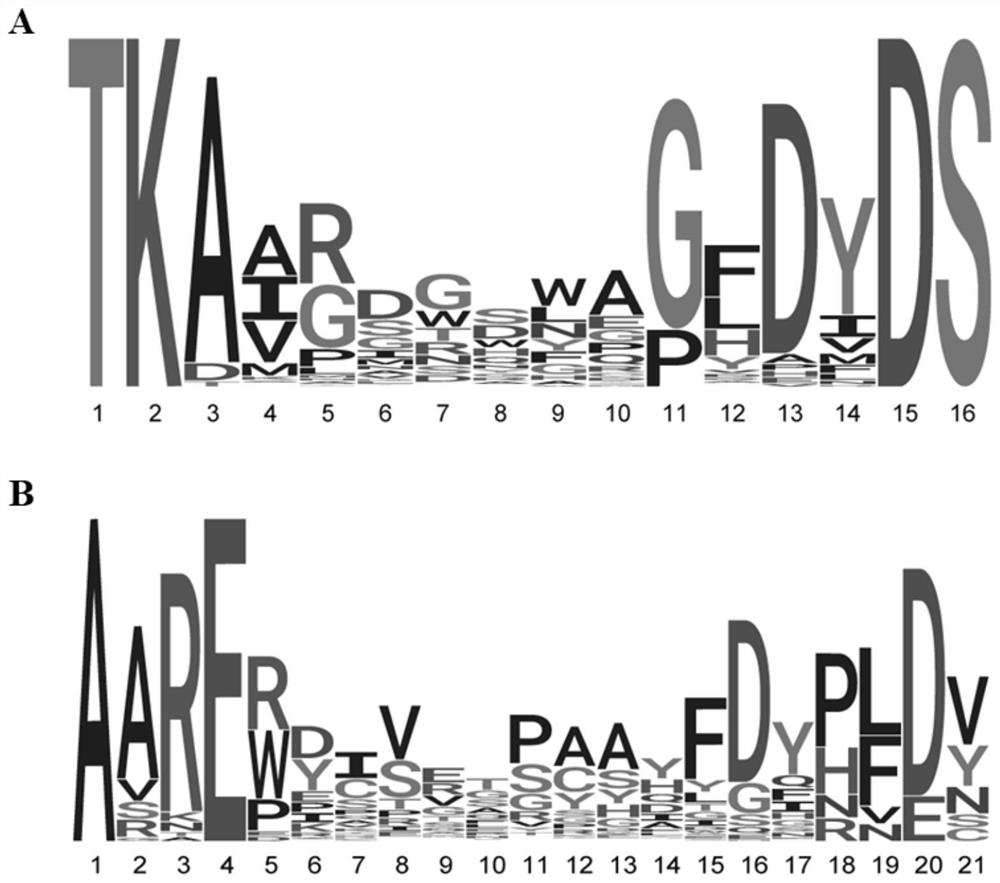
[0090] The results showed that the amino acid length of CDR3 in IgAN group was significantly shorter than that in NC group. Among them, in addition, the specific sequences of the top 10 amino acid sequences enriched in IgAN CDR3 (SEQ ID NO.37-46) and the top 10 amino acid sequences (SEQ ID NO.47-56) of the NC group are shown in Table 2 below:
[0091] Table 2
[0092] ranking IgAN panel CDR3 NC group CDR3 1 TIMES ARWDCSRTSCHQFDQ 2 ARPIGGS ARDRGRWYQLLYDPLDV 3 GGTSNGWHEIDS ARYVVLSPALGGSRLDY 4 ASGQQLGH PKIGFGYSYGGGLDV 5 ATGQQLGY AKDRSPRYDVMTGGLDS ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com