Use of branched chain alpha-keto acid dehydrogenase complex in preparing malonyl coenzyme A
A technology of malonyl coenzyme and ketoacid dehydrogenase, which is applied in the biological field and can solve problems such as low yield and biosynthesis limitation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
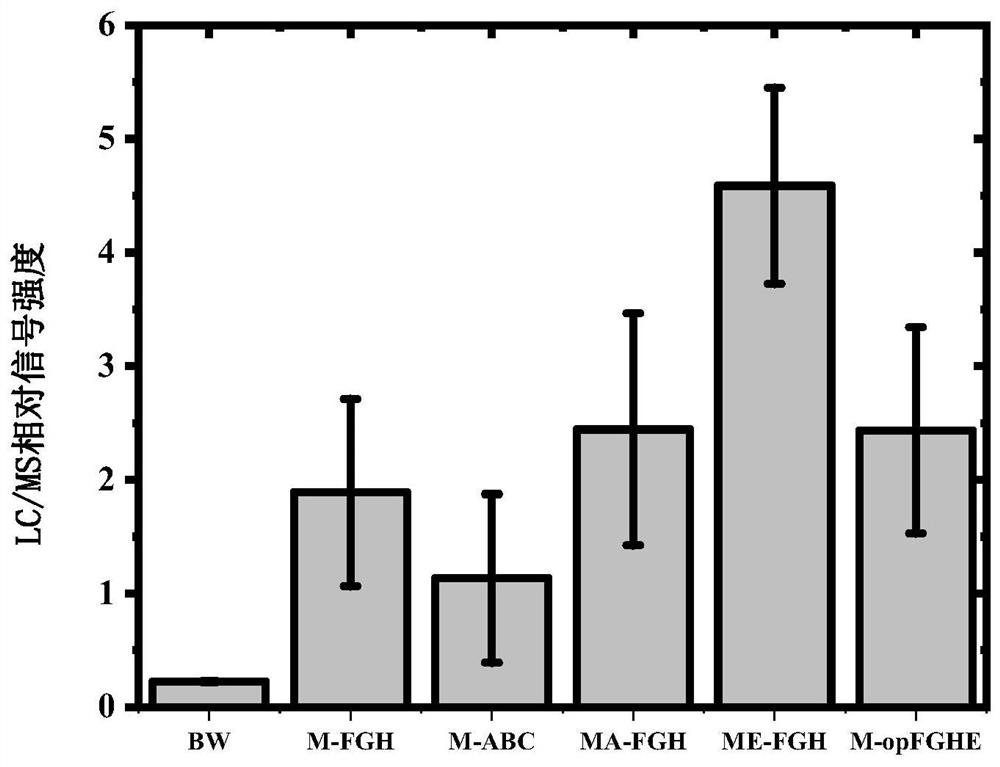
[0196] Example 1, the branched-chain α-ketoacid dehydrogenase complex can catalyze the synthesis of malonyl-CoA
[0197] The present invention finds that the branched-chain α-ketoacid dehydrogenase complex can catalyze the synthesis of malonyl-CoA, and this example prepares the coding genes (bkdA, bkdB, bkdC, lpdA1, bkdF, bkdG, bkdH genes), and further knocked out two genes (threonine deaminase ilvA and branched chain amino acid transaminase ilvE genes) in the branched-chain α-keto acid synthesis pathway. The synthesis of malonyl-CoA catalyzed by the α-ketoacid dehydrogenase complex was detected, and the primers used are shown in Table 1.
[0198] (1) Construction of a plasmid expressing the branched-chain α-ketoacid dehydrogenase complex of Streptomyces avermitilis
[0199] (1-a) PCR amplification of bkdA, bkdB, bkdC, lpdA1, bkdF, bkdG, bkdH genes
[0200] Genomic DNA of Streptomyces avermitilis was extracted using a bacterial genome extraction kit (Tiangen Biochemical Tech...
Embodiment 2
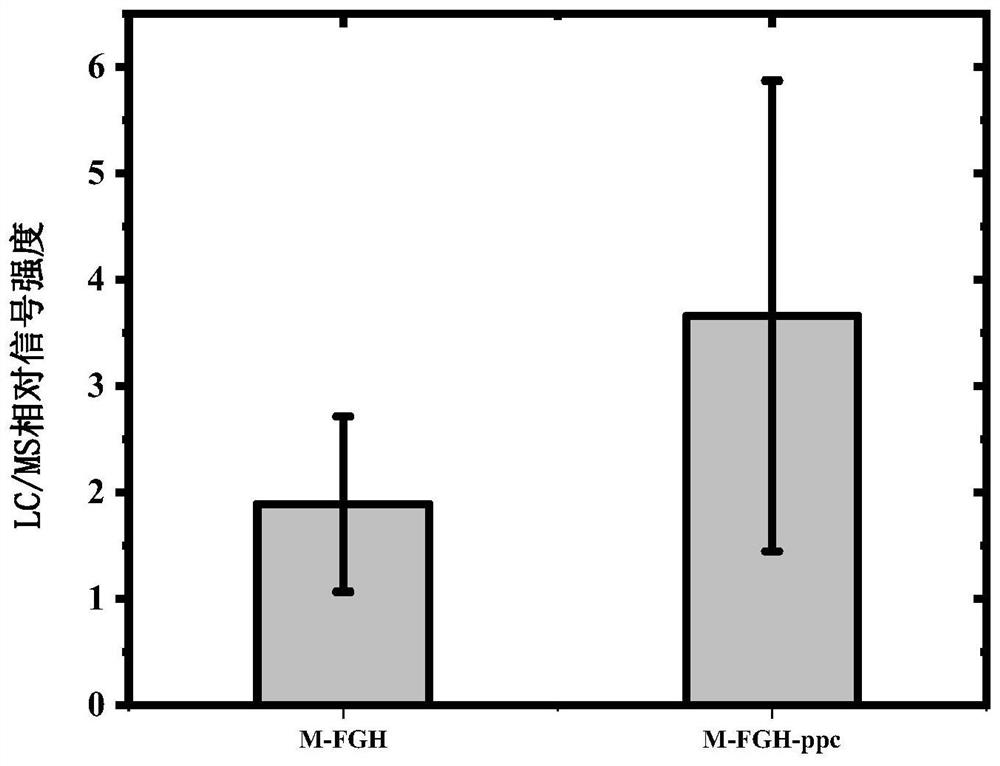
[0234] Embodiment 2, phosphoenolpyruvate carboxylase ppc gene can improve the synthesis amount of engineering strain M-FGH malonyl-CoA
[0235] In this example, on the basis of the engineering strain M-FGH in Example 1, the phosphoenolpyruvate carboxylase ppc gene was introduced and the ompT gene was knocked out (the sequence of the ompT gene is sequence 27 in the sequence listing, shown in the coding sequence 28 ompT protein), further increased the amount of synthesis of malonyl-CoA, the primers used are shown in Table 2.
[0236] (4) Construction of expressing phosphoenolpyruvate carboxylase ppc genetic engineering strain
[0237] (4-a) Genome extraction from Corynebacterium glutamicum and Escherichia coli, and PCR amplification of ppc gene, chloramphenicol resistance fragment, and upstream and downstream homology arms of ompT gene
[0238] Using ppc-F and ppc-R as PCR amplification primers, using Corynebacterium glutamicum genomic DNA as a template, amplified fragment tac-...
Embodiment 3
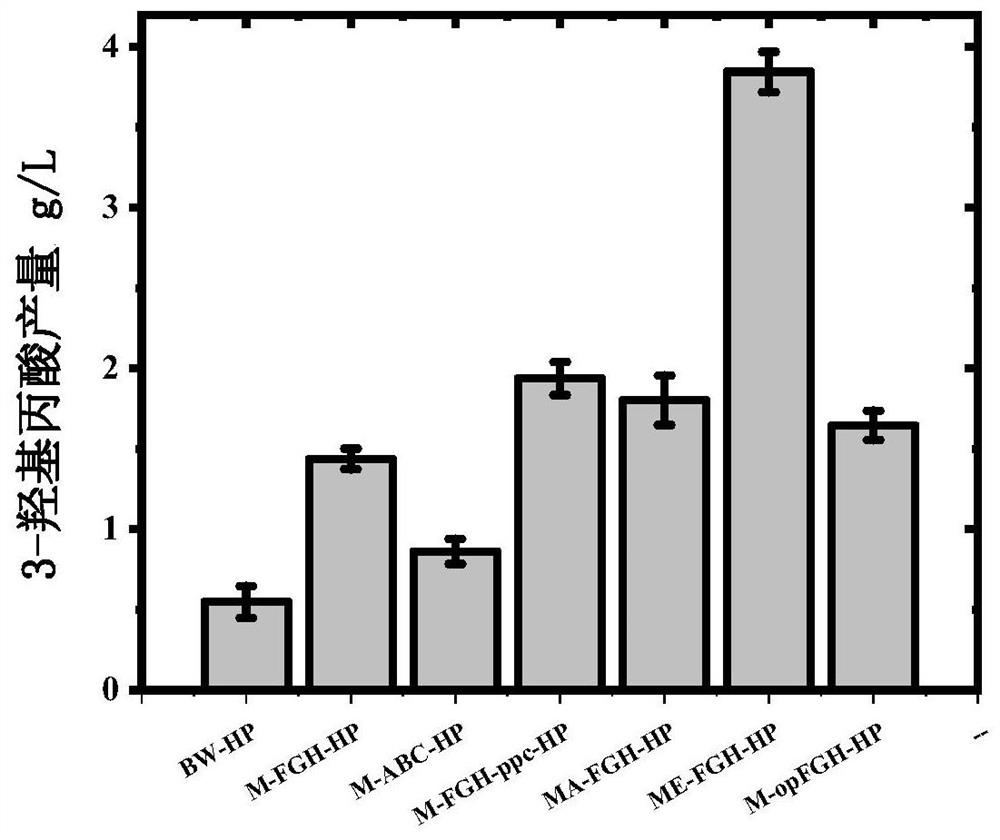
[0250] Example 3. The expression of the branched-chain α-ketoacid dehydrogenase complex gene bkdFGH-lpdA1 of Streptomyces avermitilis can increase the production of 3-hydroxypropionic acid (3-HP).
[0251] 3-hydroxypropionic acid is an important platform compound and a raw material for the synthesis of various chemicals. Malonyl-CoA can be used as a precursor to obtain 3-hydroxypropionic acid through a two-step reduction reaction. The present embodiment is by introducing thermophilic light whole Chlorothycus (Chloroflexusaurantiacus ) malonyl-CoA reductase gene mcr gene to prepare 3-hydroxypropionic acid, and use the BW of Example 1 as a control. The primers used are listed in Table 3.
[0252] (6) Construction of a plasmid expressing the malonyl-CoA reductase gene mcr of Chloroflexus aurantiacus (Chloroflexus aurantiacus)
[0253] (6-a) The nucleotide sequence of the transformed Chloroflexus aurantiacus malonyl-CoA reductase gene mcr gene is sequence 21 in the sequence list...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com