Genetically engineered bacterium for strengthening nC14-surfactin component as well as construction method and application of genetically engineered bacterium
A technology of genetically engineered bacteria and recombinant bacteria, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 Construction of Bacillus subtilis BSFX024 and BSFX025 bacterial strains
[0064] Step 1: Construct the knockout targeting fragments △pps' and △pks' according to the reverse screening marker method. Based on the principle of homologous recombination, the targeting fragments pps' and pks' were constructed by the following primers, and the pps' fragment was transformed into the competent state of BSFX022 to obtain the recombinant strain BSFX023; then the pks gene knockout fragment pks' was transformed into the strain BSFX023 In the competent state, the recombinant strain BSFX024 was obtained; then, the P veg -The bte fragment is integrated at the ackA gene locus, and the bte gene insertion fragment bte' is fused by overlapping PCR with bte series primers, and the overlapping fragment bte' is transferred into strain BSFX024 to obtain recombinant strain BSFX025.
[0065] pps-LF-F:TTTTATTTGAAAGGGAAAGGCGATCC
[0066] pps-LF-R:AATGGCCTCTGTCCGCTAATCCGCTCGGATTCCTCTCAG...
Embodiment 2
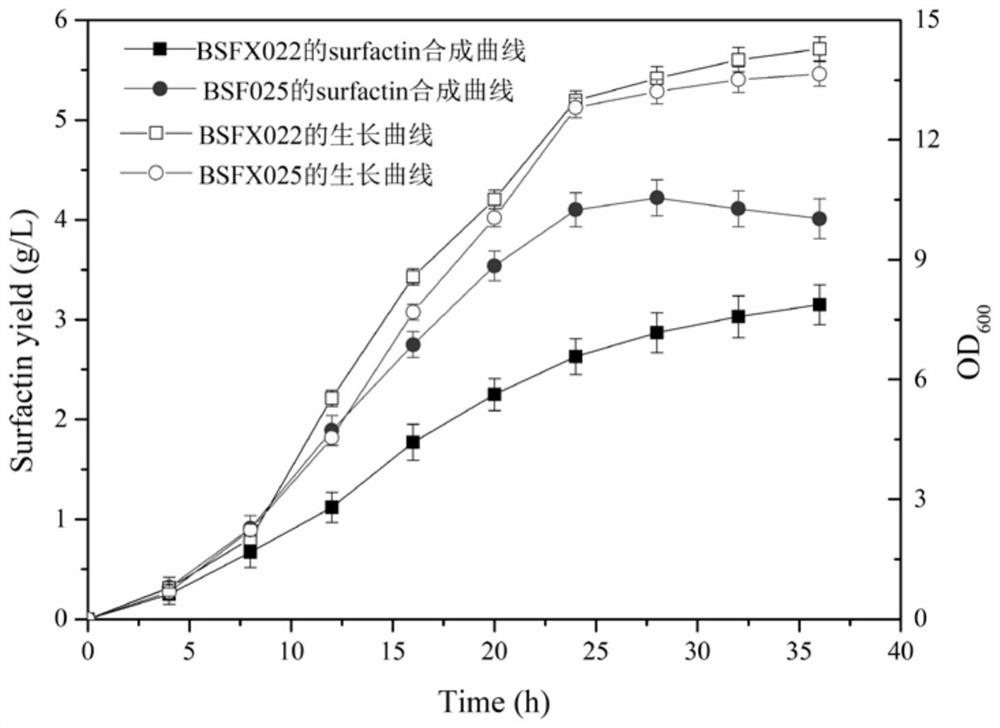
[0093] The effect of embodiment 2 Bacillus subtilis genetically engineered bacteria BSFX022 and BSFX025 strains producing surfactin
[0094] Seed medium and fermentation medium components:
[0095] The components of the seed medium are: peptone 10g / L, yeast powder extract 5g / L, sodium chloride 10g / L.
[0096] The components of the fermentation medium are: sucrose 60g / L, peptone 10g / L, sodium nitrate 6g / L, potassium dihydrogen phosphate 3g / L, disodium hydrogen phosphate 10g / L, magnesium sulfate 0.5g / L, ferrous sulfate 0.02 g / L.
[0097] The steps of producing surfactin with Bacillus subtilis genetic engineering bacteria BSFX022 and BSFX025:
[0098] (1) Seed culture medium: Add 50 mL of seed culture medium to a 250 mL Erlenmeyer flask, and sterilize at 121°C for 20 minutes. After cooling, single colonies of BSFX022 and BSFX025 were picked and placed in the seed medium at 37°C and 200 rpm for 12 hours.
[0099] (2) Fermentation culture: Add 50 mL of fermentation medium into ...
Embodiment 3
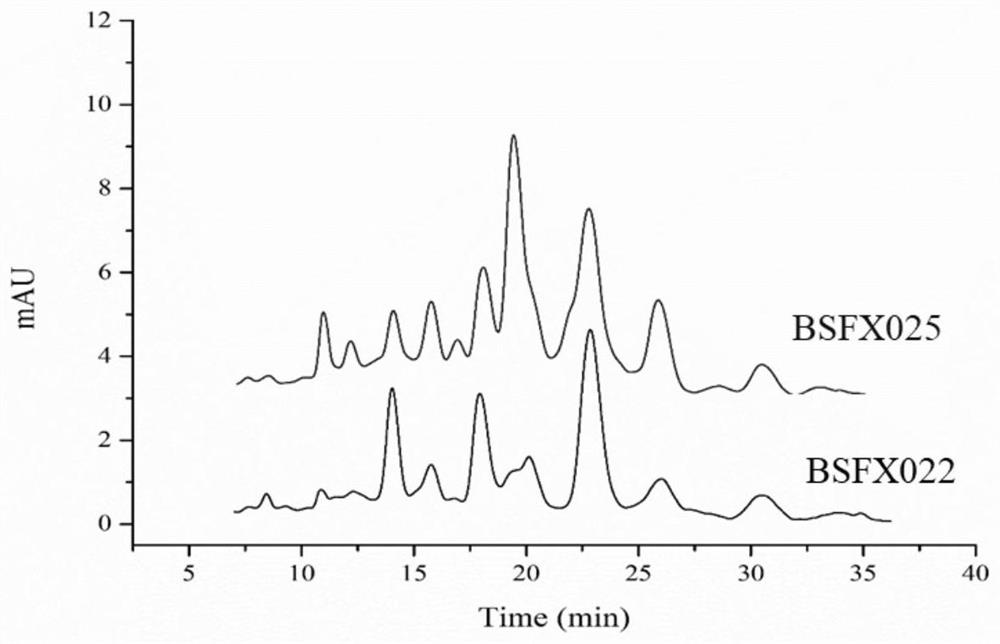
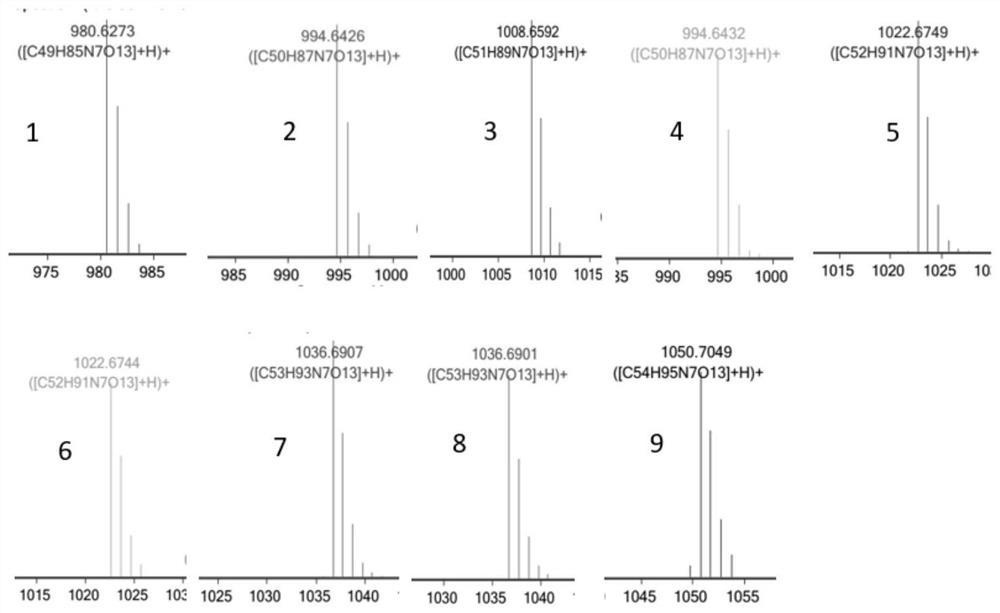
[0103] Example 3 Identification of the HPLC and HPLC-MS spectra of the surfactin crude extracts produced by strains BSFX022 and BSFX025
[0104] Firstly, the surfactin in the BSFX022 and BSFX025 strains was crudely extracted, and the extraction steps were as follows:
[0105] The first step is crude extraction of surfactin: centrifuge the fermentation broth at 8000r / min for 10min to obtain the supernatant; then use 6mol / L HCl to adjust the pH of the supernatant to 2.0, and let it stand overnight at 4°C. Centrifuge the standing liquid at 8000r / min for 10min to collect the precipitate. The pH of the precipitate was adjusted to 7.0 with 11 mol / L NaOH, and then the precipitate was freeze-dried to obtain a yellow loose solid. The yellow solid was placed in methanol and fully extracted; then the extract was dried under vacuum at 40° C. to obtain a crude surfactin product.
[0106] According to the function of bte gene, we speculate that overexpression of bte gene may produce surfa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com