Chemically modified high-stability RNA, kit and method
A highly stable, chemically modified technology, applied in biochemical equipment and methods, DNA/RNA fragments, DNA preparation, etc., can solve problems such as false negatives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
[0027] Experimental example 1 Preparation of COVID-19 RNA test template and primers
[0028] 1. COVID-19 RNA template to be tested
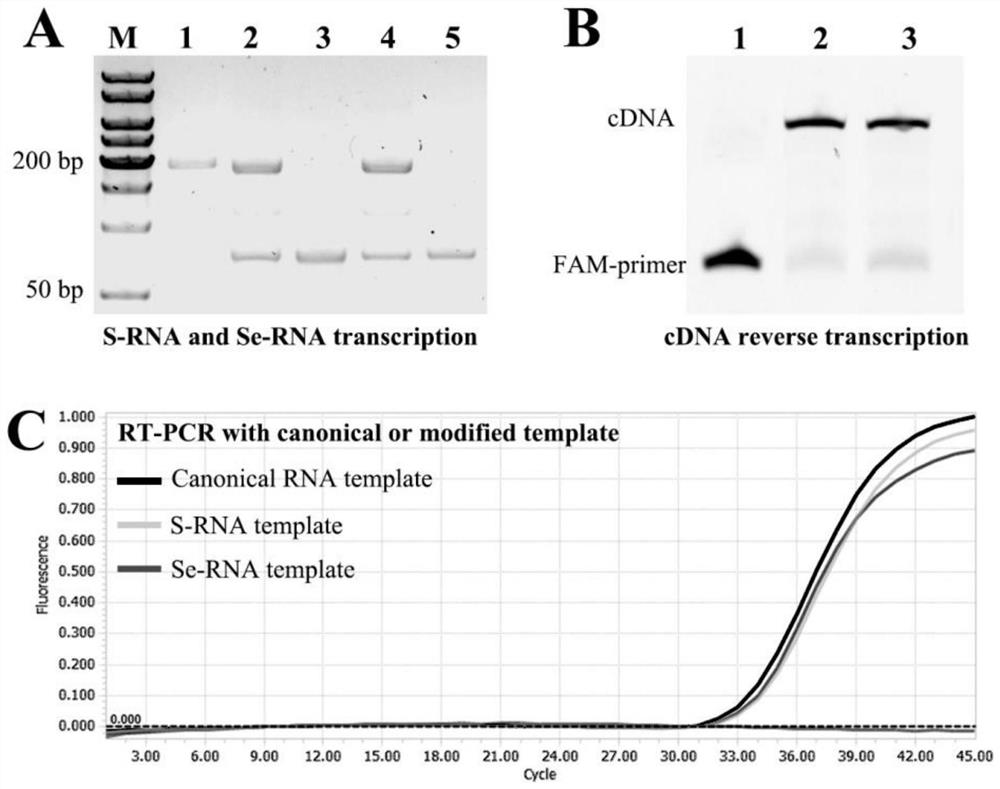
[0029] A DNA fragment with the T7 promoter and containing the COVID-19 target sequence was prepared by PCR with plasmid DNA containing the COVID-19 target sequence (pCOVID-19 plasmid; purchased from Sangon, Shanghai, China). The COVID-19 RNA (sequence shown in SEQ ID NO.1) is finally obtained through T7 transcription.
[0030] The above-mentioned COVID-19 RNA is used as a template or RNA nucleic acid to be tested for RT-PCR. Clinical COVID-19 RNA samples can also be used as templates to be tested for RT-PCR.
[0031] 2. RT-PCR primers and probes
[0032] Design forward primer F, reverse internal primer B and Taqman probe, which are finally obtained by chemical synthesis, and the specific sequences are as follows:
[0033] Forward primer F: GGGGAACTTCCTGCTAGAAT (SEQ ID NO.2);
[0034] Reverse internal primer B: GAGACATTTTGCTCTCAAGCTG (SEQ ID ...
experiment example 2
[0036]Experimental example 2 using a commercial kit to detect COVID-19
[0037] 1. Positive control of commercial RT-PCR kit
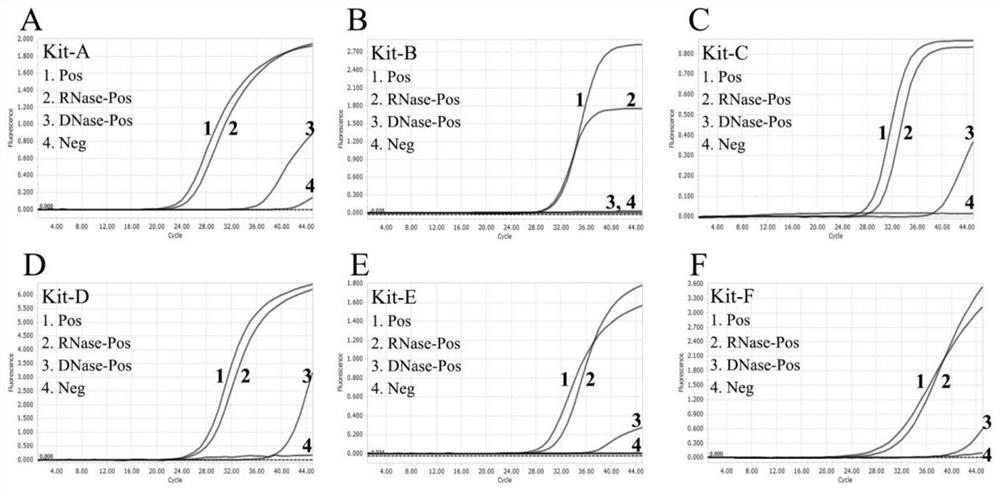
[0038] For the detection of COVID-19 RNA, 6 commercial RT-PCR kits were used for real-time RT-PCR detection, and an appropriate amount of reaction mixture was prepared according to the number of samples (COVID-19 RNA nucleic acid to be tested), positive control and negative control (including buffers, enzymes, primers and probes, included in the kit). Then, pipette template (5 μL) and reaction mixture (20 μL) into each reaction tube, then mix the reaction solution and centrifuge at low speed, and finally carry out RT-PCR amplification.
[0039] Specifically, the RT-PCR amplification reaction system is: 10X buffer buffer, 0.2mM dNTP (including dATP, dTTP, dCTP and dGTP), forward and reverse primers 0.2μM each, Taq DNA polymerase 2.5U, reverse Recording enzyme 1U, appropriate amount of RNA nucleic acid to be tested, and then add ddH 2 0 to 25 μL. Whe...
experiment example 3
[0049] Experimental example 3 commercial kit KitA detection of COVID-19 RNA
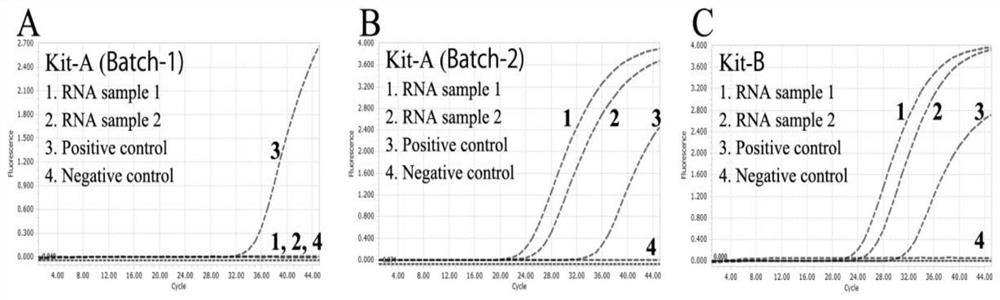
[0050] The COVID-19 virus RNA sample (COVID-19RNA) was tested using batch 1 of the commercial kit KitA (KitA, Batch-1), and its positive control DNA was used as a reference. Such as figure 2 Shown are the detection results of different batches of commercial kit Kit A and commercial kit Kit B. In each figure, curve 1: COVID-19 positive RNA sample 1; curve 2: COVID-19 positive RNA sample 2; curve 3 : the positive control of the corresponding kit; Curve 4: the negative control of the corresponding kit. As can be seen from the figure, although the DNA control reported positive results as usual, batch 1 of the commercial kit KitA failed to report positive results for these positive viral RNA samples, indicating that the reverse transcription step in RT-PCR was ineffective, but DNA aggregated and PCR reactions are running normally. The integrity of these RNA samples was confirmed by commercial kit B (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com