Cloning and functional expression method of typha orientalis heavy metal ATPase gene TyHMA3
A technology of expression method and cloning method, applied in the field of comprehensive utilization of biotechnology, can solve problems such as lack of functional expression method, and achieve the effect of improving plant stress resistance and increasing crop yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
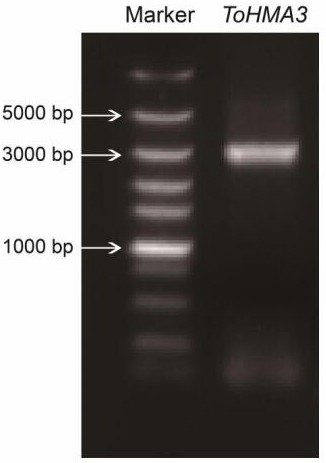
[0052] A Typha heavy metal ATPase gene THMA3 A cloning method comprising the following steps,
[0053] S1. Typha of the variety "Huaipu No. 1" was selected as the test material, and it was planted in the Laboratory of Horticultural Crop Systems Biology of Huaiyin Institute of Technology in March 2017;
[0054] S2. Put the cattail seedlings in S1 into the Hoagland nutrient solution and cultivate them. When the leaves grow to 30cm, select normal strong and consistent plants, and cut the roots, stems, pseudostems, and leaf tissue samples;
[0055] S3. Refer to the instructions of the plant RNA extraction kit from TIANGEN (Beijing) to extract the total mRNA of each sample in S2, and mix 2 μL in equal proportions;
[0056] S4. Using the PrimeScript RT-PCR Kit (TaKaRa, Japan) kit, construct a cattail cDNA library;
[0057] S5. The third-generation transcriptome sequencing data and annotation results of cattail tissue, search and splicing of cattail HMA gene;
[0058] S6. The cDNA...
Embodiment 2
[0062] A Typha heavy metal ATPase gene THMA3 The function expression method includes the following steps:
[0063] D1. Material Handling and Sampling:
[0064] (1) CK, normal Hoagland nutrient solution;
[0065] (2) T1, 0.1 mM Pb(NO 3 ) 2 deal with;
[0066] (3) T2, 0.25 mM Pb(NO 3 ) 2 deal with;
[0067] (4) T3, 0.05 mM CdCl 2 deal with;
[0068] (5) T4, 0.15 mM CdCl 2 deal with;
[0069] The root and leaf tissues were cut at 0, 2, 6, 12, 24, 48 h and 96 h, respectively, and repeated three times. Each time, 0.2 g of fresh samples were taken, quick-frozen in liquid nitrogen for 1 h, and then stored in a -80 °C refrigerator for later use;
[0070] D2. Extract the total RNA from different tissues and treatments of Typha D1, reverse transcribe it into cDNA, and detect it by real-time fluorescent quantitative PCR THMA3 relative gene expression. THMA3 Gene-specific quantitative primers qHMA3-F (TGGTTATATCAGTGTAAGAA) and qHMA3-R (CAGGAATCTATCAATCTCT), the internal re...
Embodiment 3
[0072] Example 3 THMA3 Sequence analysis
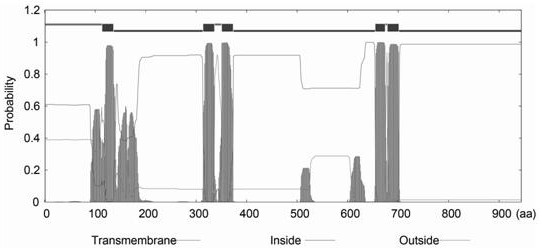
[0073] First, use the BioEdit software to find THMA3 The open reading frame (ORF) of the sequence, and translated into amino acid sequence, through NCBI (https: / / www.ncbi.nlm.nih.gov / ), Pfam (http: / / pfam.janelia.org / ) and SMART (http: / / smart.embl-heidelberg.de / ) databases to validate the obtained genes. After that, through EMBOSS (https: / / www.ebi.ac.uk / Tools / emboss / ), ProtParam
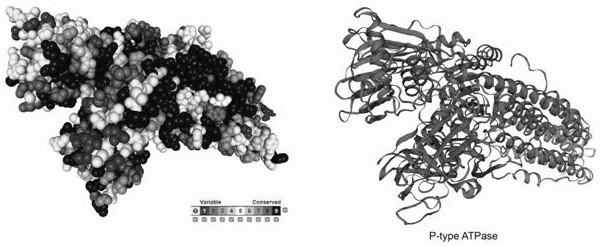
[0074] (https: / / web.expasy.org / protparam / ), TMHMM (http: / / www.cbs.dtu.dk / services / TMHMM / ), WoLF PSORT (https: / / wolfpsort.hgc.jp / ) and The SOPMA (http: / / pbil.ibcp.fr / ) program analyzes the basic characteristics of protein sequences, including molecular weight, isoelectric point, instability index, hydrophobicity, transmembrane domain and protein secondary structure. Finally, the three-dimensional protein structure analysis is predicted by ConSurf (https: / / consurf.tau.ac.il / ) and SWISS-MODEL (https: / / swissmodel.expasy.org / ), respectively.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com