Primer, probe and kit for HRAS G13R mutation detection
A mutation detection and kit technology, which is applied in DNA/RNA fragmentation, recombinant DNA technology, and the determination/inspection of microorganisms, etc., can solve the problems of sensitivity and specificity that cannot meet the detection requirements of trace samples and low cfDNA content.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
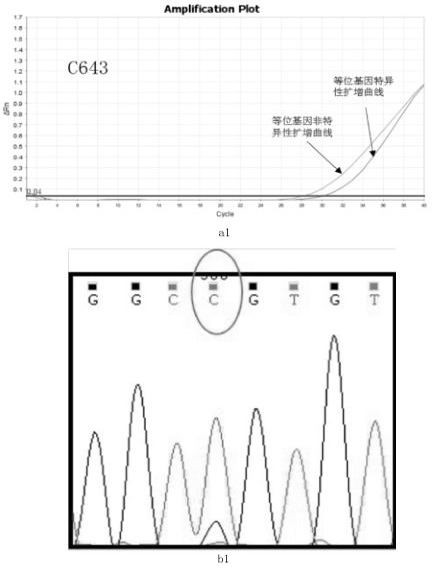
[0035] The sample to be tested is: thyroid cancer C643 cell line;
[0036] The detection method is as follows:
[0037] Step 1: Use a commercial cell / tissue DNA extraction kit to extract the genomic DNA of the cell line according to its instructions. In this example, Biotech Cell / Tissue DNA Extraction Kit (spin column type) was used to extract genomic DNA from thyroid cancer C643 cell line. The specific steps are as follows:
[0038] 1) a. Collect about 10 5 -10 6 Suspend the cells into a 1.5 ml centrifuge tube. For adherent cells, they should be digested with trypsin and collected by pipetting.
[0039] b. Centrifuge at 13,000 rpm for 10 seconds to pellet the cells. Discard the supernatant, leaving the cell pellet and about 10-20 μl residual liquid.
[0040] c. Add 200 μl buffer TB to resuspend and wash the cells, repeat the above step 1b, discard the supernatant and resuspend the cells in 200 μl buffer TB.
[0041] d. Add 200 μl of binding solution CB, immediately i...
Embodiment 2
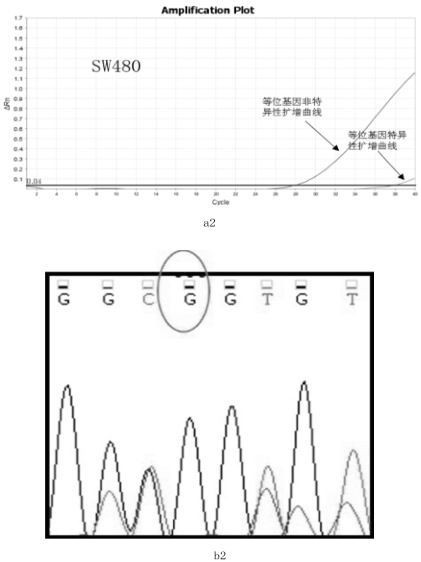
[0067] The sample to be tested is the colorectal cancer SW480 cell line;
[0068] The detection method is as follows:
[0069] Step 1: Use a commercial cell / tissue DNA extraction kit to extract the genomic DNA of the cell line according to its instructions. In this example, Biotech Cell / Tissue DNA Extraction Kit (spin column type) was used to extract the genomic DNA of the colorectal cancer SW480 cell line. The specific steps are as follows:
[0070] 1) a. Collect about 10 5 -10 6 Suspend the cells into a 1.5 ml centrifuge tube. For adherent cells, they should be digested with trypsin and collected by pipetting.
[0071] b. Centrifuge at 13,000 rpm for 10 seconds to pellet the cells. Discard the supernatant, leaving the cell pellet and about 10-20 μl residual liquid.
[0072] c. Add 200 μl buffer TB to resuspend and wash the cells, repeat the above step b, discard the supernatant and resuspend the cells in 200 μl buffer TB.
[0073] d. Add 200 μl of binding solution C...
Embodiment 3
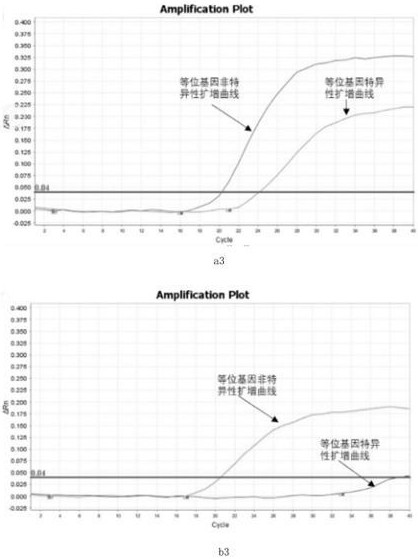
[0099] Samples to be tested: blood samples from 2 patients with thyroid cancer (i.e. blood free DNA---cfDNA)
[0100] Step 1: Use EDTA K2 anticoagulant tubes to collect blood, centrifuge at 2000-4000 rpm for 15 minutes at 4°C, and absorb the upper layer of plasma for enrichment of cfDNA. Use commercial kits or existing methods to enrich free DNA in plasma.
[0101] Step 2: Carry out the first round of PCR reaction, and use the cfDNA product obtained in Step 1 as a template to perform the following PCR reaction. The reaction system is as follows:
[0102]
[0103] Upstream primer: 5'-TCCTTGGCAGGTGGGGCA-3' (corresponding to the 7th sequence in the sequence list)
[0104] Downstream primer: 5'-CGCACTCTTGCCCACAC-3' (corresponding to the 8th sequence in the sequence list)
[0105]Probe: FAM-5'-TGAGGAGCGATGACGGAAT-3'-MGB. (corresponding to the 9th sequence in the sequence list)
[0106] The reaction conditions are as follows:
[0107] 94°C for 3 minutes; 94°C for 30 seconds,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com