O-GlcNAc glycosylation modified one-step enzyme labeling kit and application thereof
A kit and glycosylation technology, applied in the field of O-GlcNAc glycosylation modification one-step enzyme labeling kit, can solve the problems of low reproducibility and low reaction efficiency, and achieve good detection signal effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
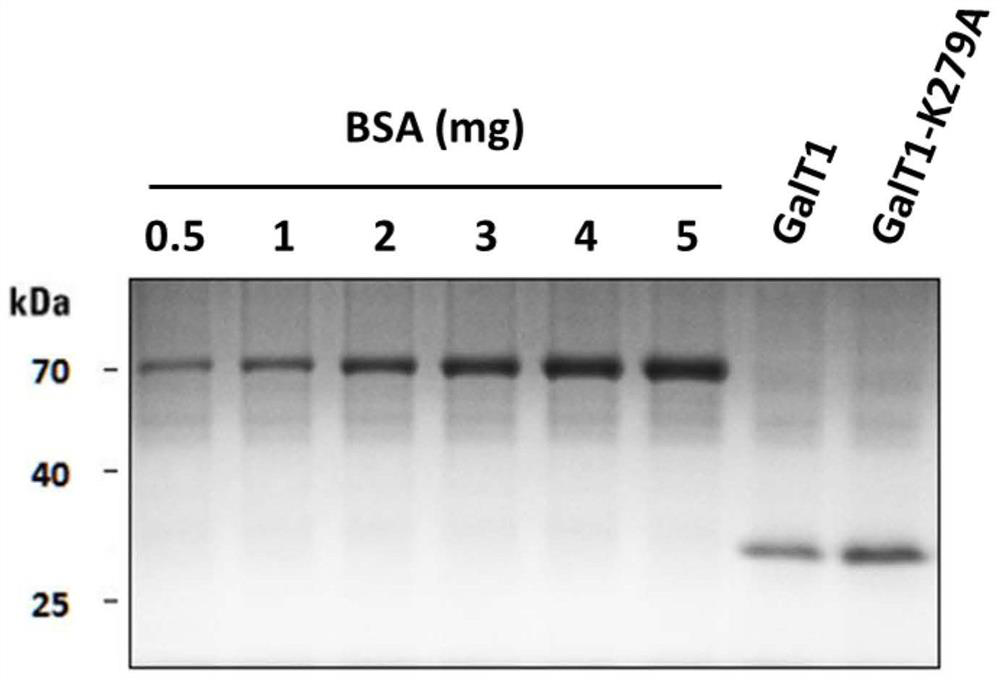
[0034] Preparation of bovine glycosyltransferase:
[0035] Expression strain construction: the company synthesized bovine glycosyltransferase GalT1 (NP_803478.1, 130-402 amino acids, pET23a, including the point mutation Y289L) gene vector, and transformed 100 μL of competent cells BL21 (DE3) with 10-50 ng of the plasmid. Place in ice bath for 30 minutes. Heat shock in a water bath at 42°C for 45 seconds, then quickly transfer the tube to ice and incubate for 2 minutes without shaking the centrifuge tube. Add 500 μL of sterile LB medium (without antibiotics) to each centrifuge tube, mix well, place at 37°C, 200 rpm, and recover for 2 hours. Paint the plate (amphicillin resistance), and incubate it upside down at 37°C for 12-24h. Pick monoclonal colonies. The GalT1-K279A protein carrier is obtained by point mutation on the basis of the GalT1 carrier. The primer sequences are:
[0036] F—AATGGATGCGTTTGGATTTAGCCTACCTTA
[0037] R—ATCCAAACGCATCCATTGCTACAGAAATGT
[0038]The p...
Embodiment 2
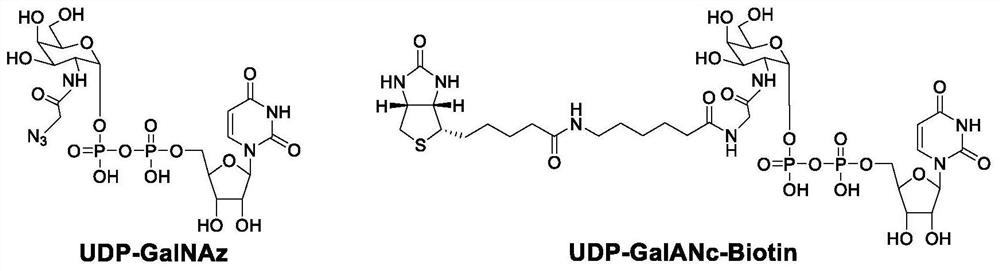
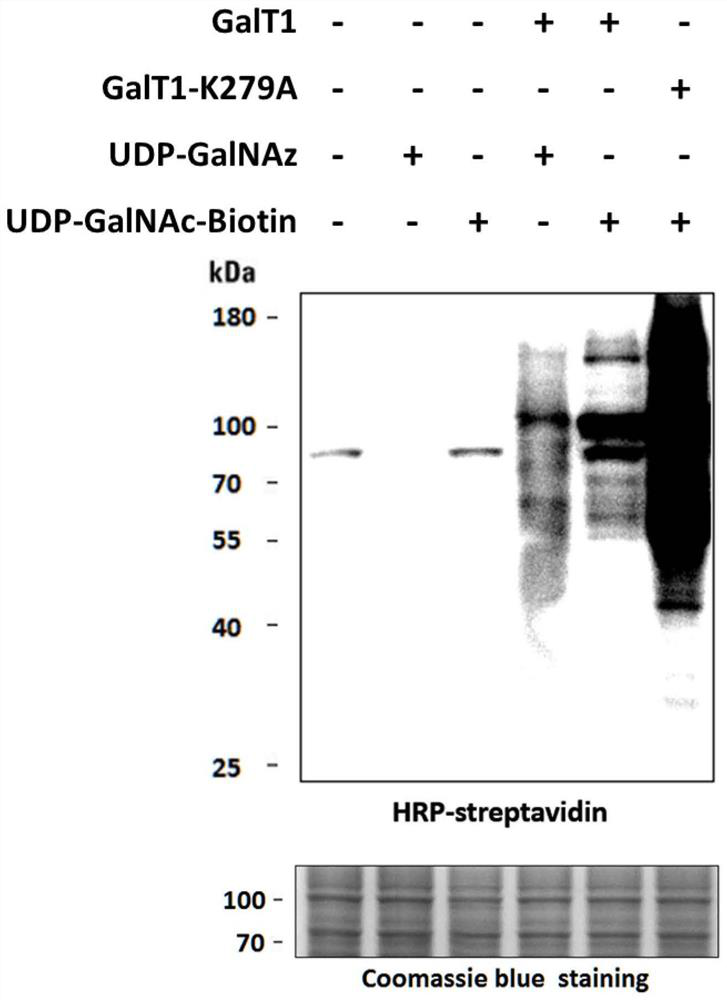
[0056] Example 2: Labeling O-GlcNAc glycosylation modification of cell lysate
[0057] (1) Collect HEK293T cells (6cm dish) by centrifugation at 4°C, 1000×g, add 300 μL of cell lysate (RIPA lysate: 50 mM Tris-HCl, pH=7.4, 150 mM NaCl, 0.1% SDS, 1×PIC protease Inhibitor mixed solution, 1mMPMSF), lysed at 4°C for 20min, centrifuged at 10000×g for 20min. The supernatant was taken, and the protein concentration was measured by BCA method.
[0058] (2) Take about 1.2 mg of protein, add 600 μL of methanol, 150 μL of dichloromethane and 400 μL of water in sequence, mix well (with white turbidity), 4 ° C, 10000 × g, centrifuge for 10 min, the white protein precipitate is in the middle, remove the upper layer solution, Add 450 μL of methanol, centrifuge to discard the supernatant, spin dry again, and evaporate the methanol to obtain a white protein precipitate.
[0059] (3) PNGase kit (NEB, P0704S) excised protein N-glycosylation modification. Dissolve the precipitated protein with ...
Embodiment 3
[0065] Example 3: Purification and omics identification of O-GlcNAc glycosylated modified protein in cell lysate
[0066] Label the cell lysate according to the operation of Example 2 (2 mg of protein per group, the reaction system is proportionally expanded), and then use streptavidin agarose resin to enrich the labeled protein, the specific operations are as follows:
[0067] (1) Take 50 μL of streptavidin agarose resin, centrifuge at 2000×g to remove the protective solution, then add 1 mL of binding buffer (100 mM Na 2 HPO 4 , 150mM NaCl, pH=7.2) wash the resin 3 times, add 1mL neutral salt solution (6% NP-40, 100mM NaCl 2 HPO 4 , 150mM NaCl): 1% SDS aqueous solution = 1:1 buffer, equilibrate the column once.
[0068] (2) For the protein that has been labeled, add 200 μL of 1% SDS aqueous solution to dissolve the protein precipitate, then add 200 μL of neutral salt solution, mix well, and centrifuge at 10000×g for 10 min. Take the supernatant, add it to the well-balance...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com