Cas9 system for efficiently editing bombyx mori genome and application of Cas9 system
A genome and silkworm technology, applied in the field of genes, can solve problems such as no baculoviruses, and achieve the effects of efficient editing, high transfection efficiency and good safety.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
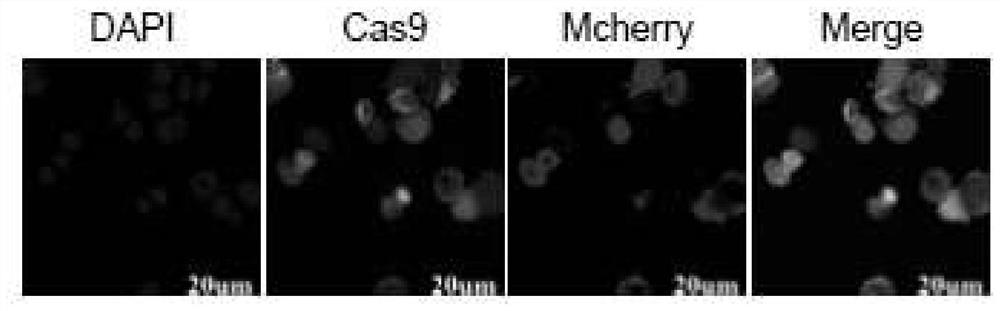
[0025] Embodiment 1, establishment of Bombyx mori Cas9 tracer cell line
[0026] Log in to the addgene vector database (http: / / www.addgene.org / ), download the existing insect Cas9 gene and U6 sequences, and design the silkworm-specific Cas9 gene and U6-sgRNA sequences according to the codon characteristics of the silkworm. The sequences are SEQID No. .1 and SEQ ID No.2, and then sent to Gensript to synthesize the sequence.
[0027] The Cas9 gene sequence was cut by Hind III and Xba I on pIZ / V5-His, and the ligation product was transformed into Escherichia coli DH5α competent cells, and then positive clones were screened with LB plates containing bleomycin, and a single positive clone was picked. The colony was used after the enzyme digestion was verified correctly, and named pIZ-OpIE2-Cas9-pA. Ligate the Mcherry gene sequence (SEQ ID No.3) to the pIZ / V5-His vector at the Hind III and Kpn I restriction sites to obtain the pIZ-OpIE2-Mcherry-pA vector, which is used after verifi...
Embodiment 2
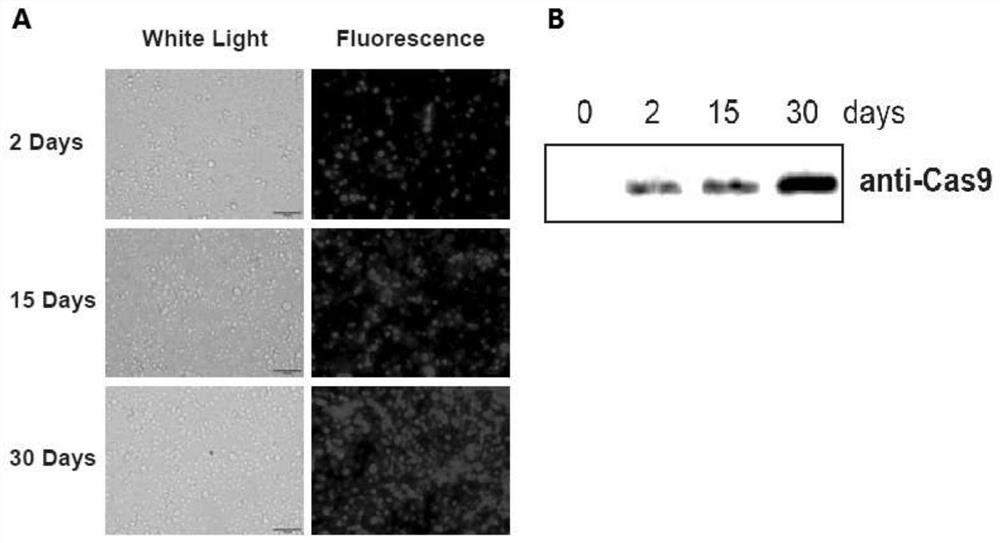
[0029] Embodiment 2, creation of Cas9 transgenic silkworm
[0030]First construct the pSL1180-IE1-Cas9-SV40 vector, digest the pSL1180-IE1-Cas9-SV40 and pBac-3Px3-EGFP vectors (SEQ ID No. 4) with AscI, and connect the excised IE1-Cas9-SV40 fragment to pBac -3Px3-EGFP vector, construct recombinant transgenic vector pBac-3Px3-EGFP-IE1-Cas9-SV40, IE1 promoter, 3Px3 promoter, EGFP gene sequences are SEQ ID No.5, SEQ ID No.6, SEQ ID No.7 shown.
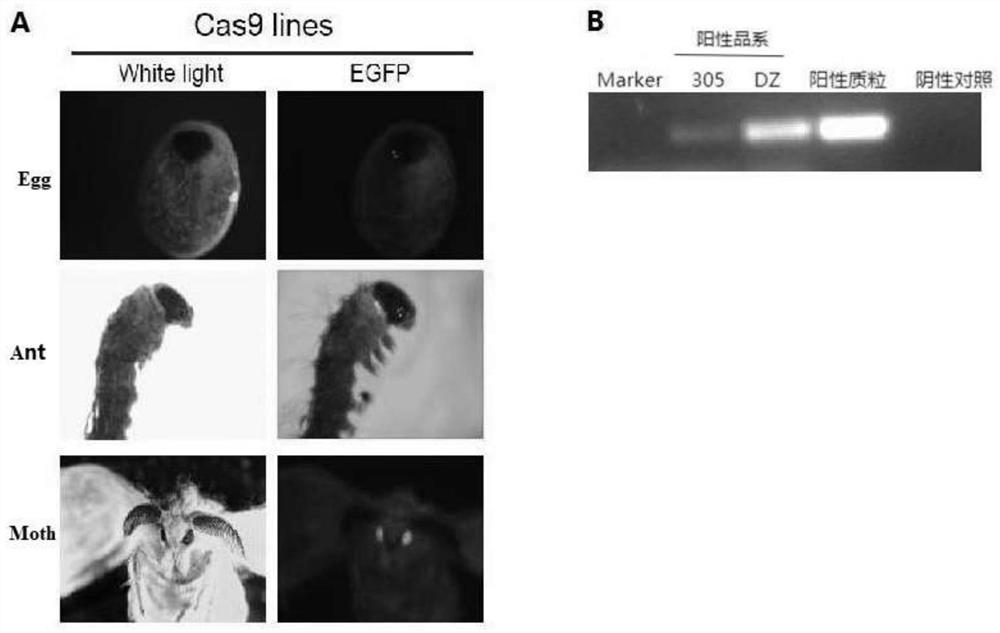
[0031] By microinjection, the pBac-3Px3-EGFP-IE1-Cas9-SV40 vector was injected into the silkworm eggs (G0 generation) within 4 hours of laying, and randomly transposed into the silkworm genome under the action of piggyBac transposon, thereby constructing Transgenic silkworm. Since 3Px3 is an eye-specific promoter, the positive transgenic silkworms were screened by observing the EGFP fluorescence in the eyes of the G1 generation silkworms. The results are as follows image 3 , shown in A. The results showed that the expression of EGFP c...
Embodiment 3
[0039] Example 3. Construction of baculovirus packaging vector
[0040] U6-sgRNA (SEQ ID NO. 2) was digested by BamHI and EcoRI and inserted into pFastBac-Dual-HSP prm - EGFP-poly vector (referred to as pFBD vector) (this vector already contains polyhedrin, which is obtained by inserting polyhedrin promoter and polyhedrin gene into pFastBac-Dual vector through SphI and KpnI), the vector sequence is as SEQ ID No.20 As shown, the recombinant vector pFastBac-Dual-HSP was constructed prm -EGFP-poly-U6-sgRNA, referred to as pFBD-U6-sgRNA vector.
[0041] The viral gene lef11 and host genes HSPD1 and ATAD3A were selected as targets to design U6-sgRNA sequences respectively. The sgRNA sequences of lef11, HSPD1, and ATAD3A were predicted according to the CRI SPRdirect online analysis tool (http: / / crispr.dbcls.jp / ), and the off-target efficiency of the target sequences in Bombyx mori was analyzed according to the software, and finally the highly efficient sgRNA sequences were selecte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com