Mercury ion detection method assisted by mimic enzyme and kit
A detection kit and detection method technology, applied in the field of biochemistry
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
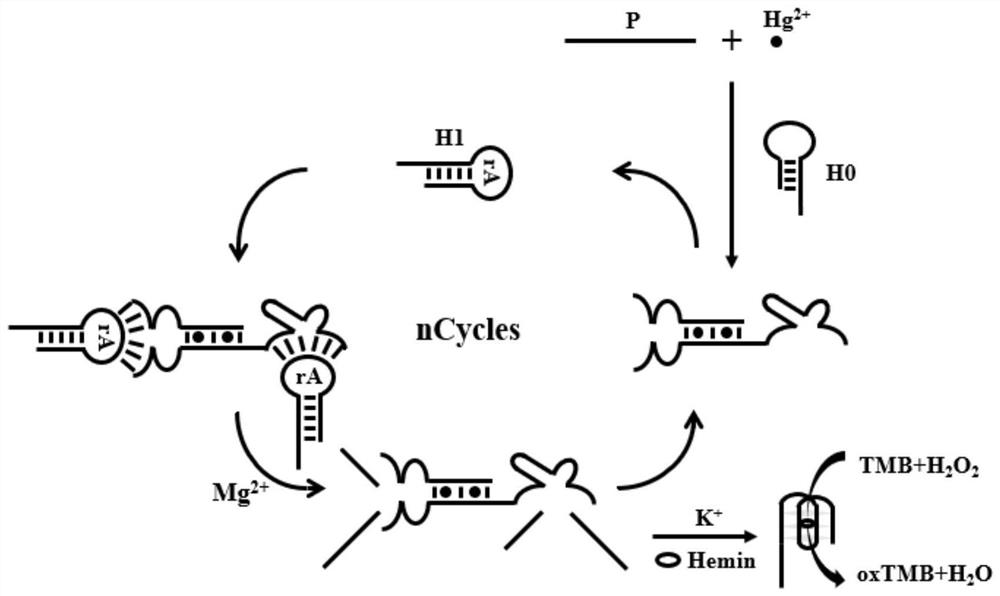
[0042] Embodiment 1: a kind of mercury ion detection method of the present invention, see figure 1 , when mercury ions are present, the auxiliary probe P and the hairpin probe H0 due to the formation of T-Hg 2+-T structure, thereby releasing the DNAzyme enclosed in the H0 loop of the hairpin probe. At the same time, the auxiliary probe P and the non-complementary part of the hairpin probe H0 form a split-type DNAzyme, and the two DNAzymes are joined by magnesium ions At the same time, it catalyzes the cleavage of the hairpin probe H1 of the modified ribonuclease (rA), releasing a large number of G-rich sequences, and the released G-rich sequences form G-quadruplexes in the presence of hemin and potassium ions Somatic DNase. The G-quadruplex DNase can catalyze H 2 o 2 The chromogenic substrate 3,3',5,5'-tetramethylbenzidine (TMB) mediated produces a blue product, which is 2 SO 4 Termination forms a stable yellow solution. The absorbance at 450 nm was measured by a UV spec...
Embodiment 2
[0047] Embodiment 2: catalytic cracking effect verification
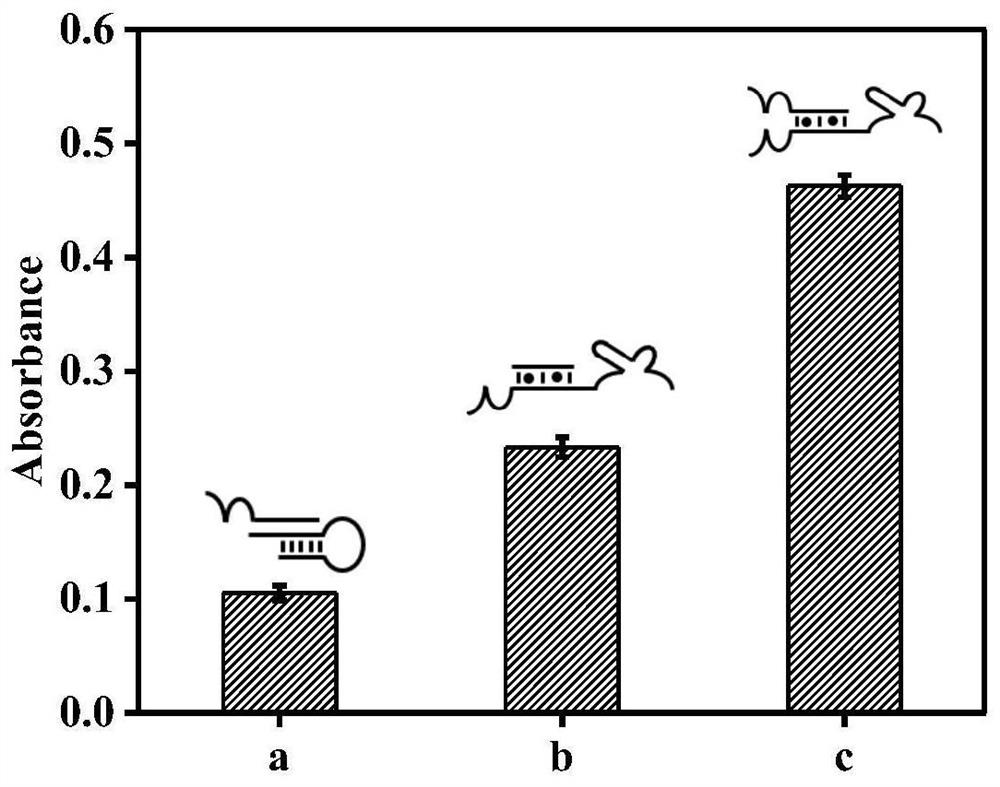
[0048] In order to verify that the double DNAzyme design has a better catalytic cleavage effect, a control probe P0 with only one DNAzyme after opening H0 was designed. The sequence of the control probe P0 is shown in Table 1 as SEQ ID No.4. The result is as figure 2 As shown, the reaction system of the double DNAzyme has greater absorbance than the single DNAzyme, and the absorbance of the two is significantly stronger than that of the blank group, indicating that the design of the double DNAzyme has a better catalytic cracking effect.
Embodiment 3
[0049] Embodiment 3: Hairpin probe H1 and H0 concentration ratio optimization
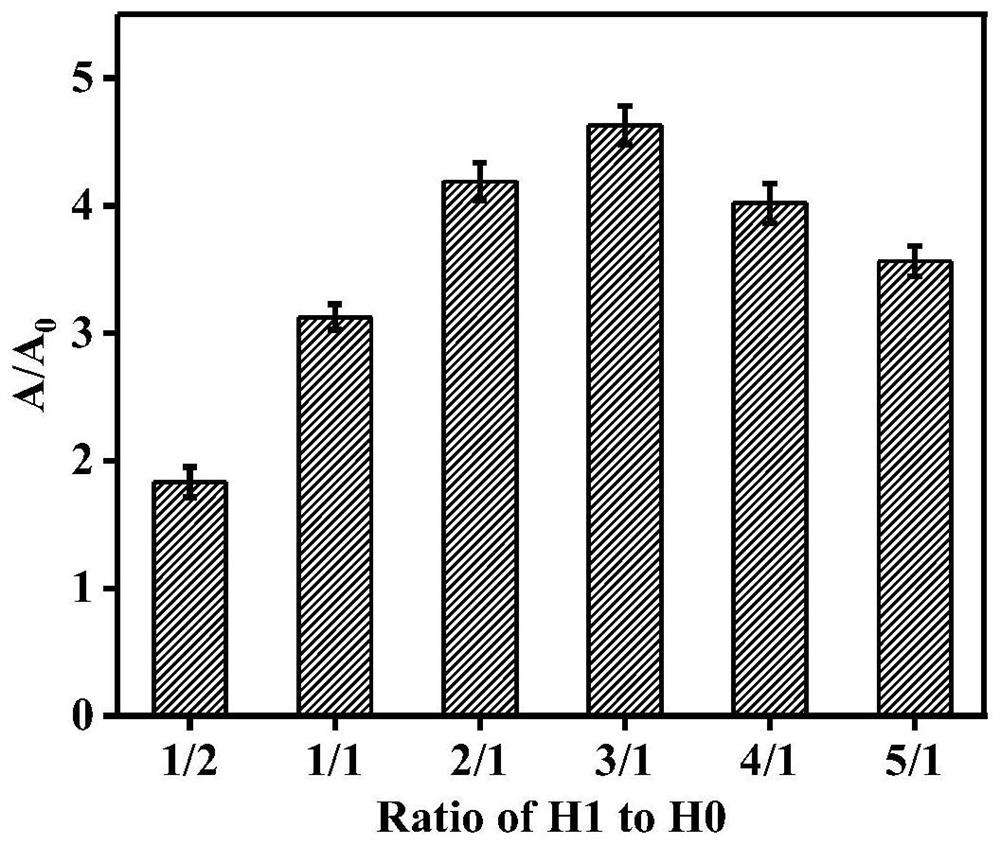
[0050] In order to obtain the best experimental conditions, the effect of the concentration ratio of hairpin probes H1 and H0 on the absorbance was studied. The concentration of fixed auxiliary probe P and hairpin probe H0 was 20nM, and 10, 20, 40, 60, 80, 100nM hairpin probe H1 was added to the detection system respectively. According to the steps in Example 1, other conditions were kept constant. Change the concentration of hairpin probe H1 to optimize. The result is as image 3 As shown, the effect is best when the concentration of hairpin probe H1 is 60 nM. Therefore, the concentration ratio of hairpin probes H1 and H0 was selected as 3:1 as the optimal concentration ratio.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com