Recombinant plasmid combination, genetically modified saccharomycetes and method for producing odd-chain fatty acid
A technology for recombining plasmids and fatty acids, which is applied in the field of genetically modified yeast and the production of odd-chain fatty acids. It can solve the problems of high fermentation costs, difficulties in the production and ratio of odd-chain fatty acids, and achieve low fermentation costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0171] Construction of embodiment 1 expression plasmid
[0172] (1) Construction of propionyl-CoA synthetase (prpE) expression plasmid Y33-prpE
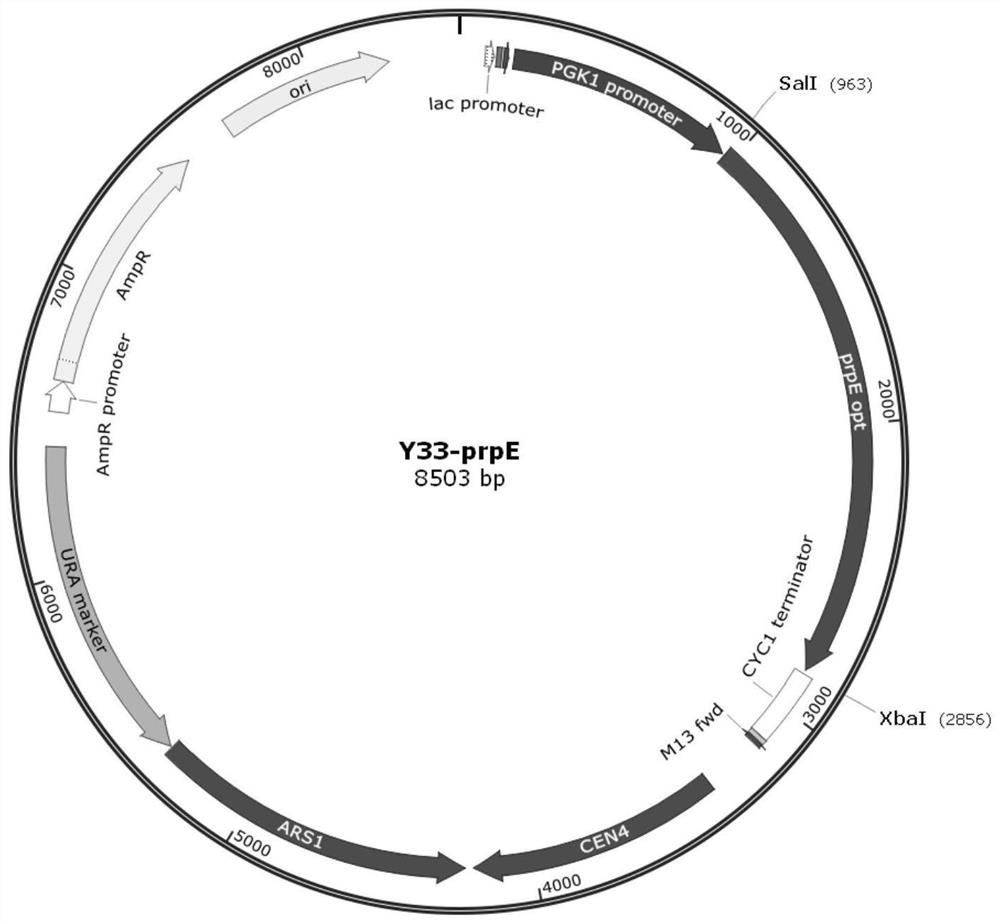
[0173] The polynucleotide sequence (prpE) of propionyl-CoA synthetase of Salmonella enterica was selected for codon optimization, and the optimized polynucleotide sequence is shown in SEQ ID NO.1. After cutting the nucleotide sequence shown in SEQ ID NO.1 with SalI and XbaI, insert it into the vector Y33-PGKCYC (shown in SEQ ID NO.2) between SalI and XbaI restriction sites to obtain propionyl-CoA synthesis Enzyme expression plasmid Y33-prpE., its plasmid map is as follows figure 1 shown.
[0174] (2) Threonine deaminase (tdcB) expression plasmid Y22-tdcB construction
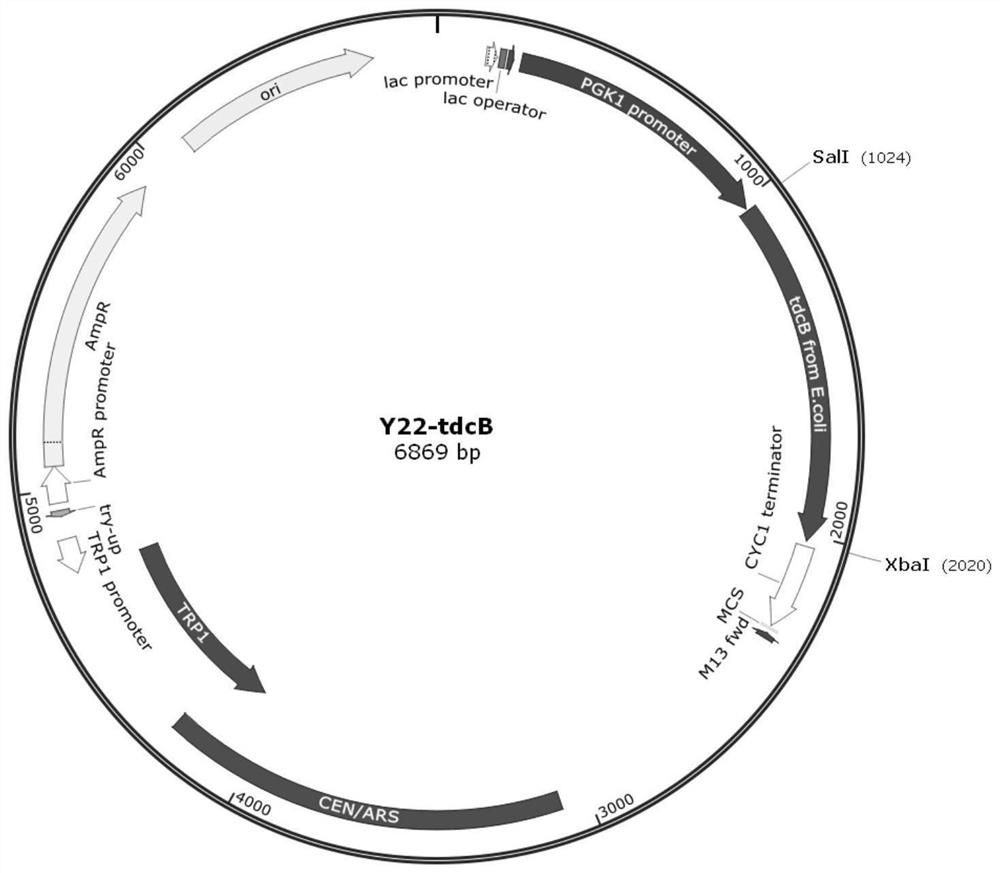
[0175] The threonine deaminase tdcB gene (NC_000913.3 (3265039..3266028), shown in SEQ ID NO.3) from Escherichia coli was selected for expression. Using the Escherichia coli genome as a template, polymerase chain reaction (PCR) was carried out with the following p...
Embodiment 2
[0214] Example 2 Construction of Aspartokinase, Homoserine Dehydrogenase and Threonine Synthetase Co-expression Plasmid Y33-thrABC
[0215] Aspartokinase, homoserine dehydrogenase and threonine synthase were all selected from Escherichia coli, and were composed of thrA(NC_000913.3(337..2799), thrB(NC_000913.3(2801..3733) and thrC (NC_000913.3 (3734..5020) encoding.
[0216] a. First construct the thrA expression plasmid
[0217] 1) Using the Escherichia coli genome as a template, carry out a PCR reaction with the following primers:
[0218] Primer ThrA-F (5'-3'):
[0219] TTATCTACTTTTTCACAAAATATAAAACAGTCGACATGCGAGTGTTGAAGTTCGGC;
[0220] Primer ThrA-R (5'-3'):
[0221] TGACATAACTAATTACATGATGCGGCCCTCTAGATCAGACTCCTAACTTCCATGAGAGG;
[0222] Table 13. PCR reaction system:
[0223] Element Volume (microliter) upstream primer 2.5 downstream primer 2.5 Template (E. coli genome) 0.5 5×Q5 Reaction Buffer 10 dNTPs (10mM) 1 Q5 DNA poly...
Embodiment 3
[0287] Example 3 Construction of aspartate kinase, aspartate aminotransferase and phosphoenolpyruvate carboxylase co-expression plasmid Y33-HOM3mu-ppc-aspC
[0288] Aspartate kinase is from Saccharomyces cerevisiae, encoded by HOM3 gene (NC_001137.3(256375..257958)); aspartate aminotransferase is from Escherichia coli, encoded by aspC gene (NC_000913.3(984519..985709)); Phosphoenolpyruvate carboxylase is from Escherichia coli and is encoded by the ppc gene (NC_000913.3(4150447..4153098)).
[0289] a. Construction of HOM3 expression plasmid Y33-HOM3mu
[0290] 1) Using the Saccharomyces cerevisiae genome as a template, carry out a PCR reaction with the following primers:
[0291] Primer HOM3-F (5'-3'):
[0292] CTACTTTTTTACAACAAATATAAAAACAGTCGACATGCCAATGGATTTCCAACCTA;
[0293] Primer HOM3-R (5'-3'):
[0294] TGACATAACTAATTACATGATGCGGCCCTCTAGATTAAATTCCAAGTCTTTTCAATTGT;
[0295] Table 25 PCR reaction system:
[0296] Element Volume (microliter) upstream pri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com