Ultrahigh-flux single cell sequencing method
A single-cell sequencing and cell technology, applied in the field of single-cell sequencing, can solve the problems of not easy portability, high sequencing cost, and expensive equipment.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] 1. Microplate preparation
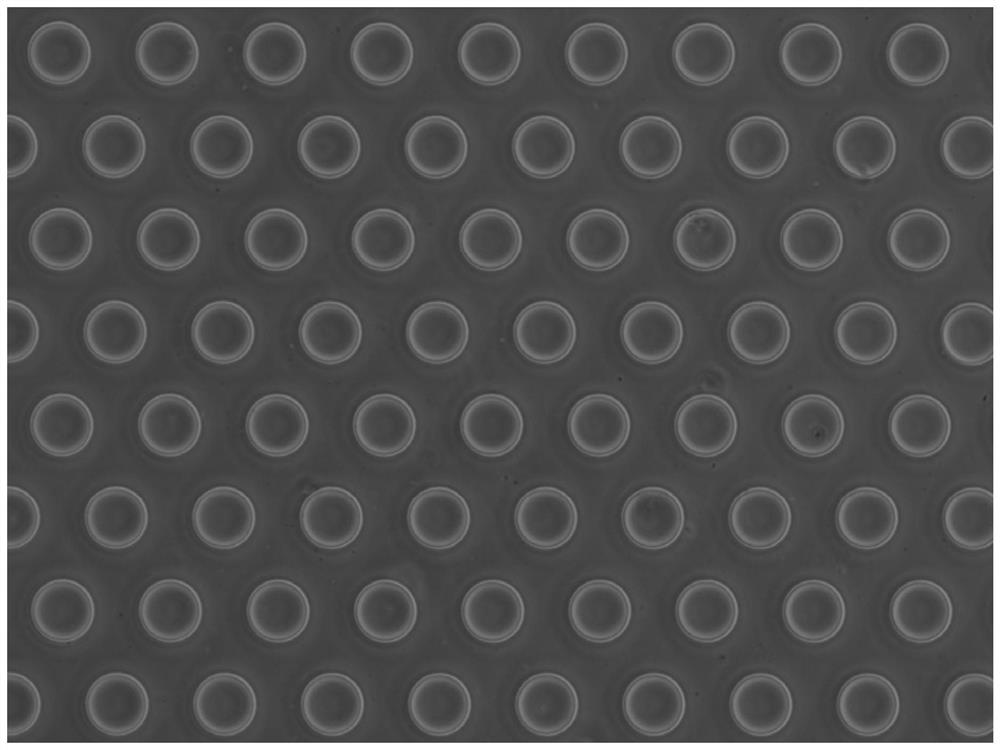
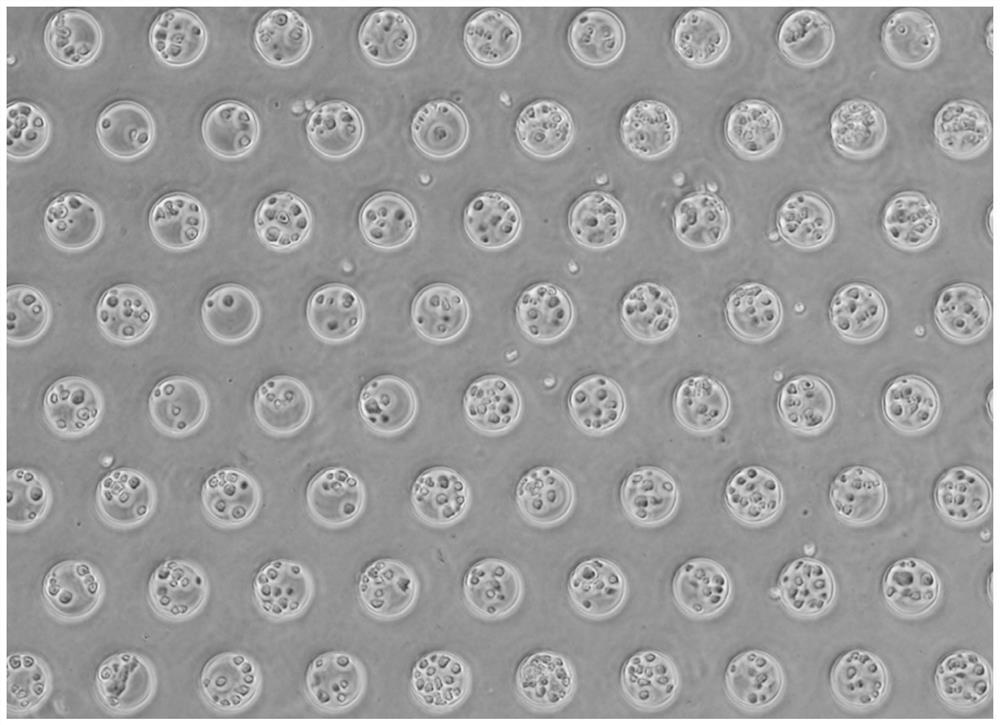
[0056] According to the experimental scale (500,000 human 293T cells and 500,000 mouse 3T3 cells each), design the size of the microwell plate (the size of the well plate is 1.8cm×1.8cm), and etch the microwell on the silicon wafer as the initial mold, and the microwell is a cylinder Body shape, in which the depth of micropores is 60 μm, the diameter of micropores is 50 μm, and the distance between pores is 70 μm. Next pour polydimethylsiloxane (PDMS) on the silicon chip, take off PDMS after molding and become the second mold that has microcolumn on the plate, the microporous plate that final experiment uses is that concentration is 5% (mass ratio ) agarose (prepared with enzyme-free water), poured on the PDMS microcolumn plate after hot melting to condense and form, and the agarose plate at this time is a microporous plate with a certain thickness after being peeled off ( figure 1 ). When saving, add DPBS-EDTA mixture which is harmless to...
Embodiment 2
[0067] Human 293T, mouse 3T3 mixed cell test.
[0068] Add 5 million mouse embryonic stem cells (ESC) 3T3 and human embryonic kidney cells (293T) each 5 million slowly dropwise with 5-10ml methanol (-20°C pre-cooled) respectively, fix at -20°C for 30 minutes, and at the same time, aliquot the bridging primers 6.5 μl per well into eight tubes, then dispensed into a 96-well plate containing 0.5 μl reverse transcription primers, and left to mix to form a total of 1 μl reverse transcription mixed primers per well. The bridging primer sequence is 5'- CGTCGTGTAGGGAAAGAGTGT GACGCTGCCGACGA[ddC]-3', the 3' end is modified with ddC to prevent the extension of the bridging primer during reverse transcription and lead to the generation of by-products.
[0069] There are also 96 kinds of reverse transcription primers (reverse transcription sequences) and the above-mentioned cell label sequences, and the core sequence is 6×N. Each primer is placed independently in each well. This 6×N rand...
Embodiment 3
[0085] cDNA sequencing library construction.
[0086] (1) 5ng starting DNA fragmentation
[0087] Vazyme TD512 kit was used.
[0088] (a) Thaw 5×TTBL (TruePrep Tagment Buffer L) at room temperature, invert and mix well before use. Confirm that 5×TS (Terminate Solution, reaction termination solution) is at room temperature, and flick the tube wall to confirm whether there is precipitation. If there is precipitation, heat at 37°C and shake vigorously to mix well and the precipitation will dissolve.
[0089] (b) Add each reaction component in sequence in a sterilized PCR tube:
[0090] 5 × TTBL 4μl
[0091] DNA 5ng
[0092] TTE Mix V1 5μl
[0093] wxya 2 O to make up to 20 μl.
[0094] (c) Use a pipette to gently pipette 20 times to mix thoroughly.
[0095] (d) Put the PCR tube in the PCR instrument, and set the following reaction program: 55°C for 10 minutes; keep warm at 10°C.
[0096] (e) Immediately add 5 μl 5×TS to the reaction product, and mix well by gently blowi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Pore depth | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com