Nucleic acid aptamer binding to novel coronavirus (SARS-CoV-2) nucleocapsid protein and use thereof
A nucleic acid aptamer, nucleocapsid protein technology, applied in the direction of viruses/phages, viruses, viral peptides, etc., can solve the problems of renal failure, rapid spread, and imperfect detection methods for new coronaviruses, and achieves small molecular weight, Chemically stable, easy to store and label effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1: Screening of ssDNA nucleic acid aptamers specifically binding to novel coronavirus (SARS-CoV-2) nucleocapsid protein
[0046] 1. Synthesize the random single-stranded DNA library and primers shown in the following sequences:
[0047] Random ssDNA library:
[0048] SEQ ID NO: 5: 5'-GTTCGTGGTGTGCTGGATGTNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTGACACATCCAGCAGCACGA-3'
[0049] Among them, 36 "N"s represent a sequence formed by connecting 36 arbitrary nucleotide bases. The library was synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
[0050] The primer information is shown in Table 1 and was synthesized by Nanjing GenScript Biotechnology Co., Ltd.
[0051] Table 1 Primers and their sequences
[0052]
[0053] Among them, S in the primer name represents the forward primer, A in the primer name represents the reverse primer, 19 A in the sequence represent a polyA tail composed of 19 adenine nucleotides (A), and "Spacer 18" represents an 18-atom Hexaethy...
Embodiment 2
[0076] Embodiment 2: Surface plasmon resonance (SPR) detects affinity and specificity detection of SARS-CoV-2 nucleocapsid protein nucleic acid aptamer and SARS-CoV-2 nucleocapsid protein
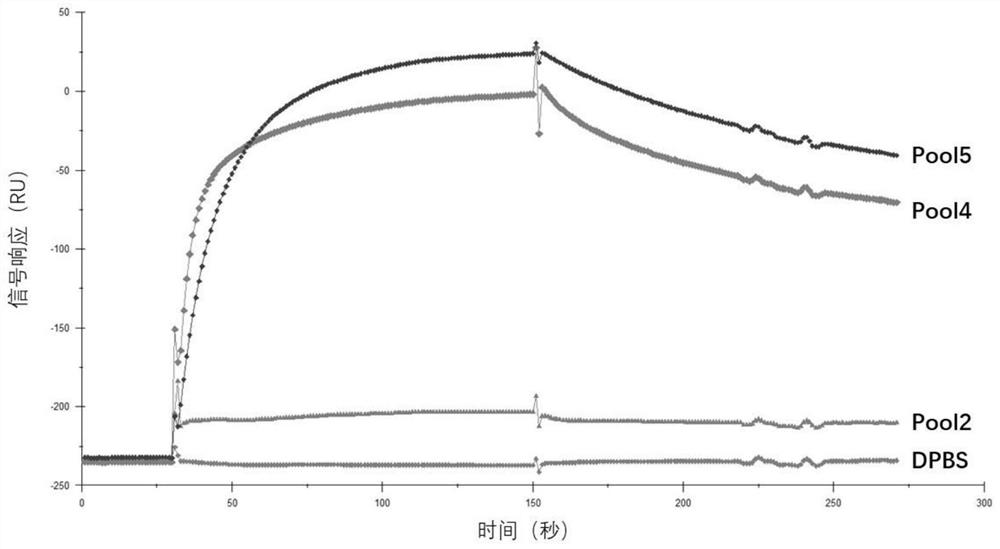
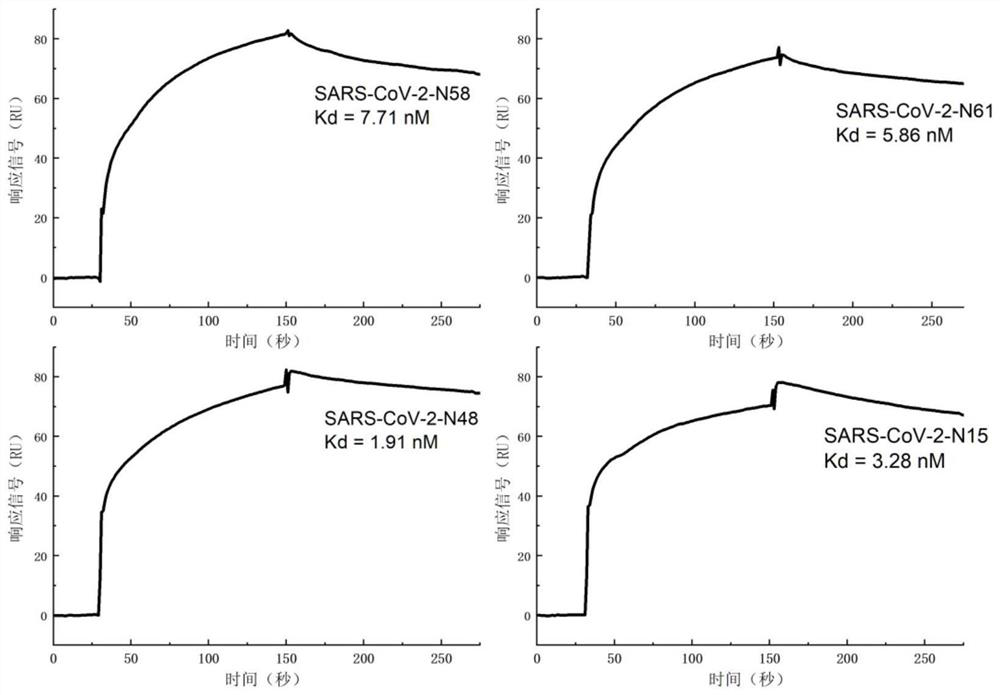
[0077] 1. Dilute the nucleic acid aptamers SARS-CoV-2-N15, SARS-CoV-2-N48, SARS-CoV-2-N58, and SARS-CoV-2-N61 of general biosynthesis with DPBS buffer respectively into 500nM.
[0078] 2. Coupling the SARS-CoV-2 nucleocapsid protein to the second channel on the surface of the CM5 chip: first wash the chip with 50mM NaOH, inject 20 μl of the sample at a flow rate of 10 μl / min, and then use an equal volume of EDC (1-( 3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride; 0.4M aqueous solution) and NHS (N-hydroxysuccinimide; 0.1M aqueous solution) were mixed and injected into 50 μl activated chip , with a flow rate of 5 μl / min. The SARS-CoV-2 nucleocapsid protein was diluted with 10 mM sodium acetate at pH 5.5 to a final concentration of 50 μg / mL and then injected. The injection volume w...
Embodiment 3
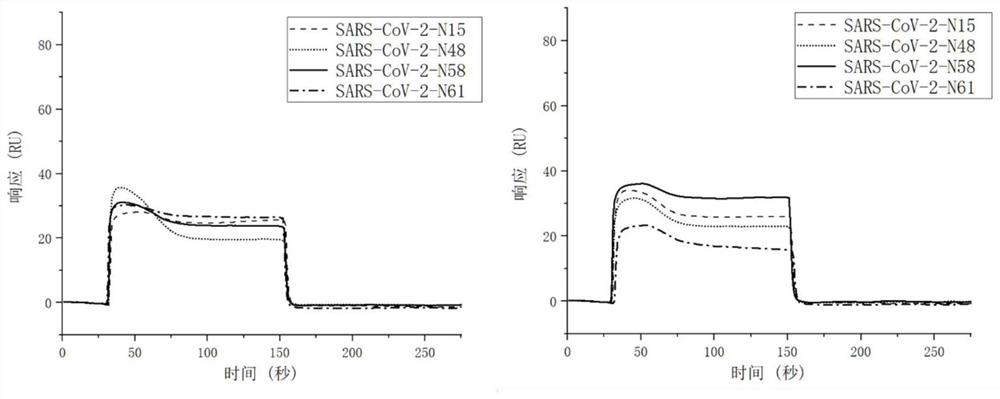
[0085] Example 3 shows the binding of the four nucleic acid aptamers screened in Example 1 of the present invention to the binding site of the N protein using surface plasmon resonance (SPR)
[0086] Using surface plasmon resonance (SPR), using the chip of Example 2, the four nucleic acid aptamers of general biosynthesis were diluted to 500nM respectively, and SARS-CoV-2-N48 was injected for 2 minutes, and then SARS-CoV- 2-N58 injection for 2 minutes, dissociation for 2 minutes. Such as Figure 4 As shown, it can be seen that the signals of the two nucleic acid aptamer samples were significantly improved, which can explain that SARS-CoV-2-N48 and SARS-CoV-2-N58 bind to different sites of the N protein. The same experimental method verified that SARS-CoV-2-N48 binds to SARS-CoV-2-N61 at different sites on the N protein, and SARS-CoV-2-N15 binds to SARS-CoV-2-N58 on the N protein SARS-CoV-2-N15 and SARS-CoV-2-N61 bind to different sites of the N protein. SARS-CoV-2-N48 and SA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com