Application of circular RNA as osteoarthritis marker
An osteoarthritis and annular technology, applied in the field of biomedicine, can solve problems such as cartilage degeneration of regenerated cartilage tissue, and achieve the effect of significant significance and good application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
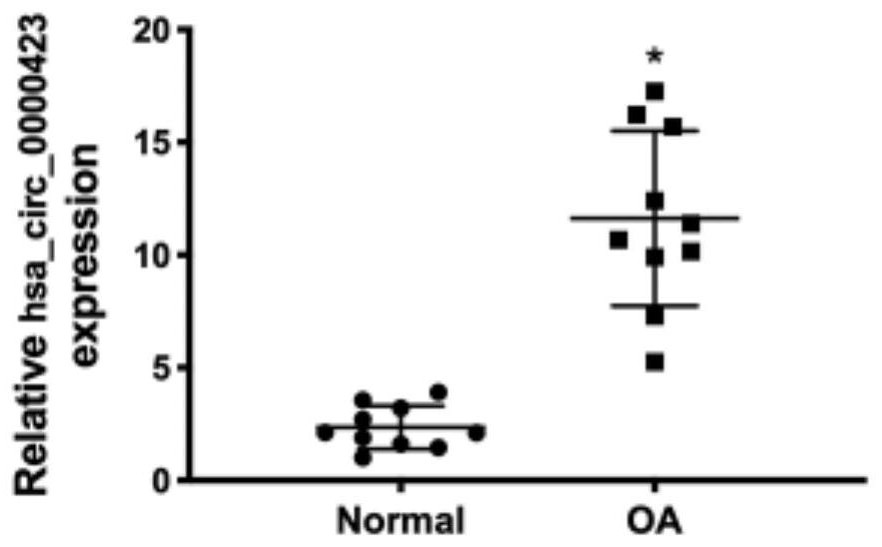
[0046] This example studies the expression of circular RNAhsa_circ_0000423 in normal volunteers and patients with osteoarthritis.
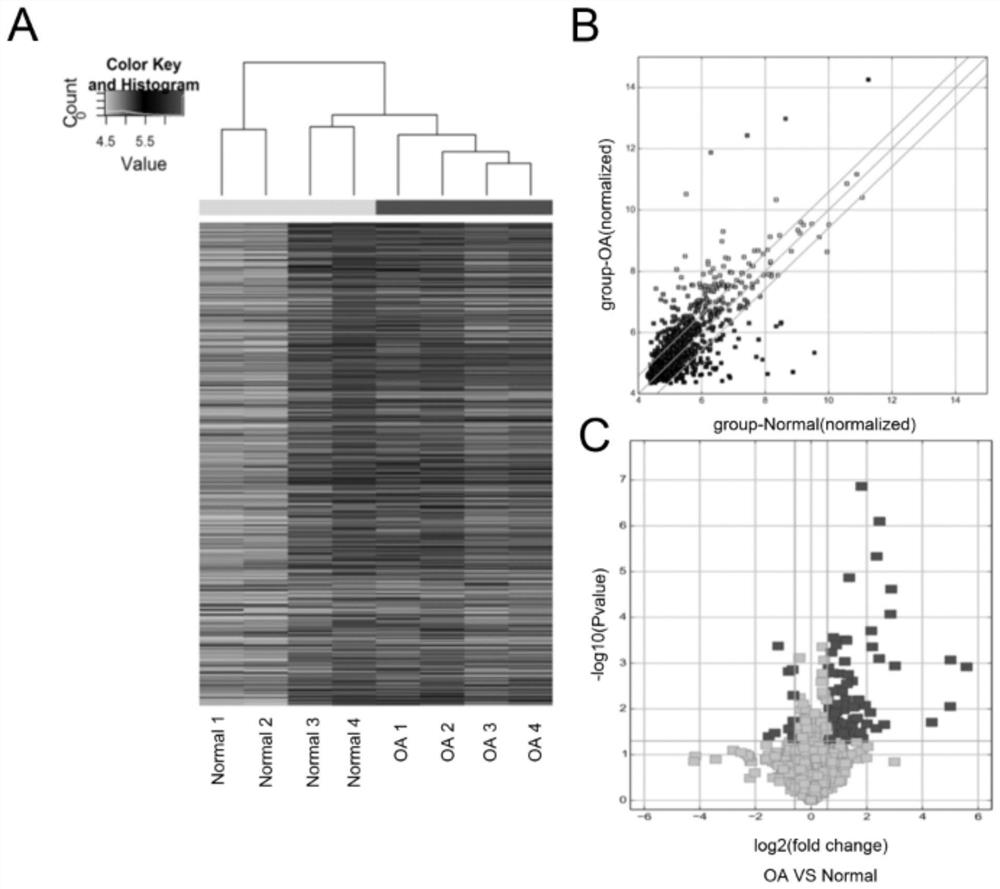
[0047] Using Array Human Circular RNA gene chip sequencing to detect 4 cases of cartilage tissue samples from normal (Normal) and osteoarthritis (OA) patients, and conducted bioinformatics analysis, the expression profiles of circRNAs that were found to be different in the samples were found. The results are as follows: figure 1 shown. in, figure 1 Figure A in the middle is the clustering diagram of the expression profile of circRNAs, which shows that there are significant differences in the expression profiles of circRNAs between the two groups, and the expression profiles in each group are relatively consistent; figure 1 Figure B in the middle is a scatter plot of circRNAs, and the lines outside the lines are circRNAs with expression difference > 2; figure 1 Middle C is the volcano map of differentially expressed circRNAs, and the dark dots sh...
Embodiment 2
[0049] 1. Biological characteristics of circular RNAhsa_circ_0000423
[0050] Circular RNA hsa_circ_0000423 is used as an OA marker, including 1138bp, and its full nucleic acid sequence is shown in SEQ INNO.1.
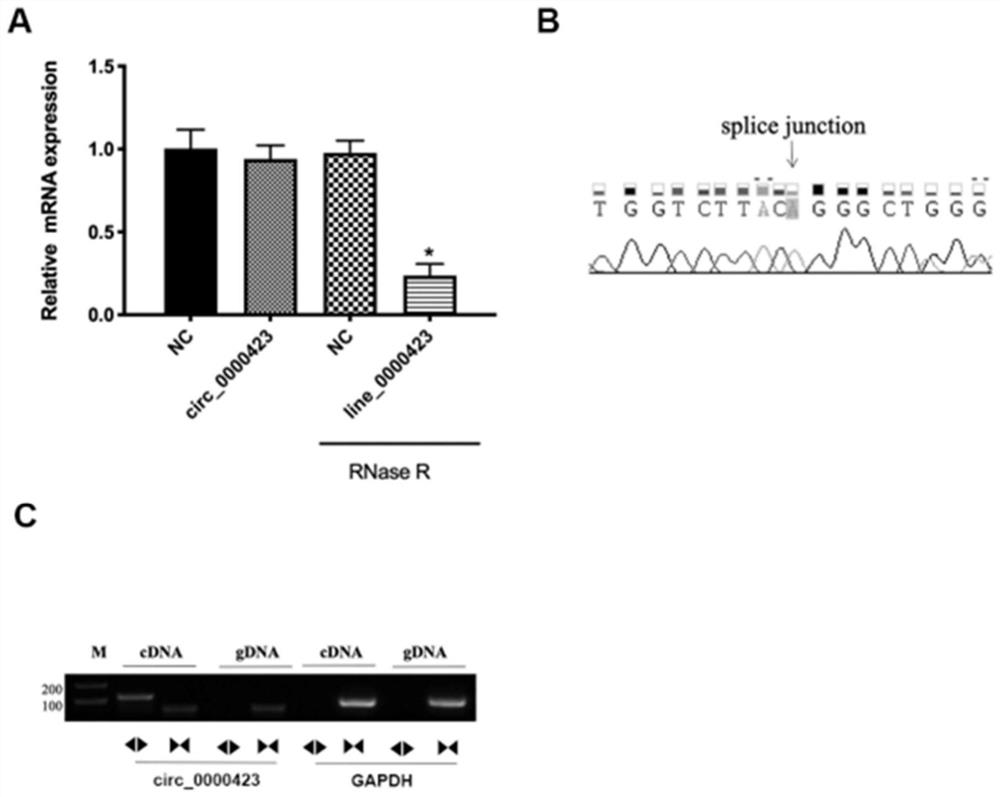
[0051] 2. Circular verification of circular RNAhsa_circ_0000423
[0052] The specific primer of the OA marker based on circular RNAhsa_circ_0000423: including the forward primer of hsa_circ_0000423 and the reverse primer of hsa_circ_0000423, the nucleotide sequence of the forward primer of hsa_circ_0000423 is shown in SEQ IN NO: 2, the reverse of hsa_circ_0000423 The nucleotide sequence of the primer is shown in SEQ INNO:3.
[0053] RNase R enzyme digestion: use GENESEED RNase R digestive enzyme, configure 10 μL of reaction system according to 2 μg total RNA + 0.5 μL RNase R + 1 μL Reaction Buffer, and use RNase free H for insufficient volume 2 O supplementation, 37°C, incubation for 20 minutes, reverse transcription at 70°C for 10 minutes to inactivate the enzyme, a...
Embodiment 3
[0063] The effect of hsa_circ_0000423 on the biological function of OA was verified by transfecting the overexpressed hsa_circ_0000423 plasmid, which was provided by Shanghai Heyuan Biotechnology Co., Ltd.
[0064] By transfecting the overexpressed hsa_circ_0000423 plasmid into human chondrocytes, Western Blot technology was used to detect the protein levels of related metabolic products (MMP13 and COL-2) in human chondrocytes. The specific steps are as follows:
[0065] 1. Transfection
[0066] 1. One hour before transfection, suck off the medium in the 6-well plate, replace with 2mL opti-MEM medium per well or the serum-free medium corresponding to the cells, and place at 37°C, 5% CO 2 in a constant temperature incubator.
[0067] 2. According to the number of transfected plasmids, prepare enough EP tubes, and clearly mark the name of the plasmid on the tube cap. One of the EP tubes is the main tube of opti-MEM medium, and one EP tube is the PEI transfection reagent (or Tra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com