Visual rapid nucleic acid detection method of bovine nodular skin disease virus
A detection method and nodularity technology, which is applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve the problems of high cost, insensitive detection method, long time, etc., and achieve specificity High efficiency, accurate and reliable detection results, and low detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
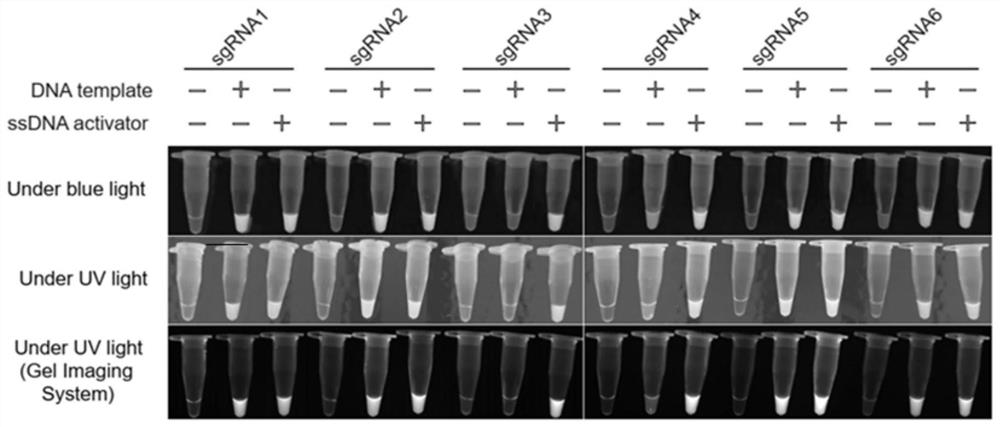
[0071] Example 1: Screening of sgRNA for bovine nodular skin disease gene visualization detection
[0072] 1.1 Determination of target genes
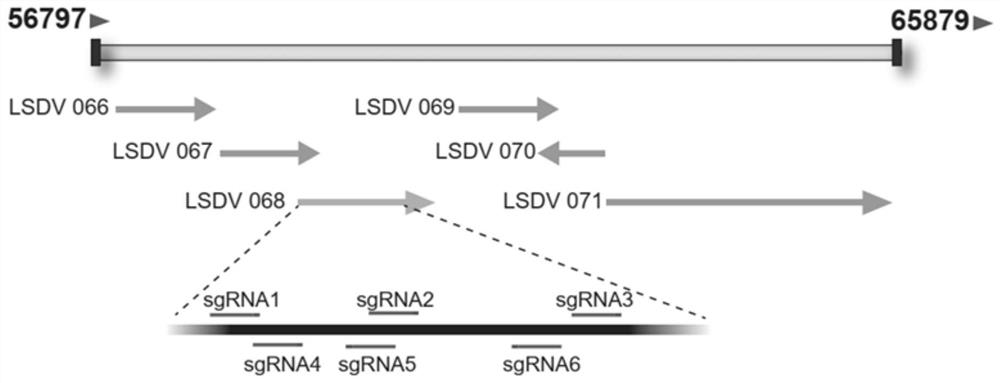
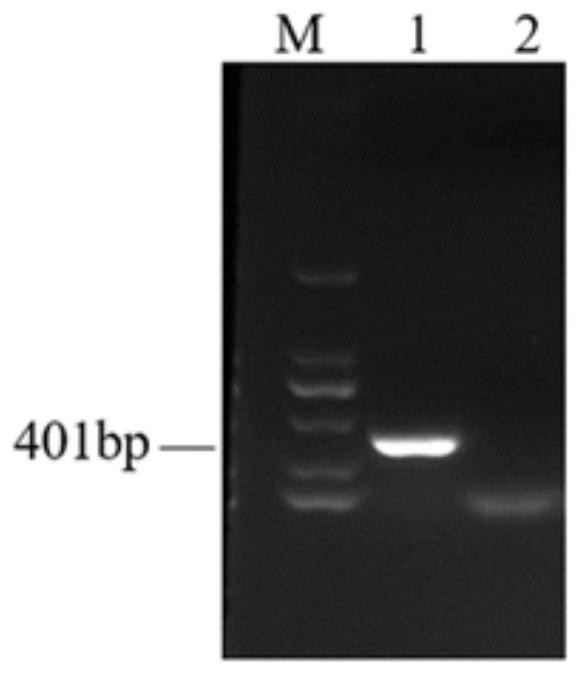
[0073] Using LSDV / Russia / Saratov / 2017 (GenBank: MH646674.1) as the standard, compared all the viruses in the genus of capipoxvirus in NCBI, and found the most conserved gene sequence. The gene name in LSDV is LSDV orf068 , within the scope of the gene, search for the PAM sequence, and find that there are 6 sgRNAs within the range of 401bp, the sequences are shown in SEQ ID NO:1-6, and their positions in the genome are as follows figure 1 shown.
[0074] The fragment was amplified with the sequence shown in PCR-F / R (SEQ ID NO:7, SEQ ID NO:8), the amplification program was: 98°C, 5min; 35 cycles (98°C, 10s; 58°C, 15s; 72°C, 30s) 72°C, 5min. The amplification system is: PremixSTAR Max Premix (TAKARA product number: R045A): 25 μL; PCR-F / R: 2.5 μL each; DNA template: 1 μL; double distilled water to make up 50 μL. The result is as figur...
Embodiment 2
[0086] Embodiment 2: Screening of RPA primers
[0087] The 401bp fragment in Example 1.1 was constructed into the pUC57 plasmid and named pUC57-orf068. According to the formula: copy number = (concentration × 6.022 × 10 23 ) / (full length of plasmid×1×10 9 ×650) to calculate the copy number, pUC57-orf068 was diluted to different copy numbers as the template for RPA reaction. According to the RPA primer design principle, 4 pairs of primers (SEQ ID NO.18-25) were designed for the orf068 gene, and the amplified product contained sgRNA1. RPA amplification was performed for each copy number of plasmids using different primer pairs. The RPA reaction configuration is as follows: Primer free Buffer: 29.5 μL; DEPCwater: 12.2 μL; RPA-F / R. Mix well and add to a TwistAmp Basic reaction to fully dissolve, then add 2.5μL of 280mM magnesium acetate to each system. Finally, 1 μL of plasmid DNA of each copy number was added, and reacted at 37° C. for 40 minutes. Take 3 μL of the RPA produ...
Embodiment 3
[0095] Embodiment 3: This detection method compares with qPCR detection sensitivity
[0096] 1. Detection sensitivity to plasmids
[0097] Each Cas12a digestion reaction system of the second pair of primers in Example 2 was transferred to a black 96-well microtiter plate, then 80 μL of DEPC water was added, and after mixing, the fluorescence value was measured with a multifunctional microplate reader. Each reaction was repeated in triplicate. The result is as Figure 8 . It shows that the detection sensitivity can also reach 5 copies / μL, which is consistent with the result of naked eye observation in Example 2. The plasmid diluted in Example 2 was used as a template to verify the sensitivity of qPCR. The detection target of this qPCR method is also the LSDVorf068 gene, and the RPA amplification region completely includes the qPCR amplification region. The detection results of qPCR primers (SEQ ID NO:26-27) are shown in Table 1. It is found that the sensitivity of qPCR is a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com