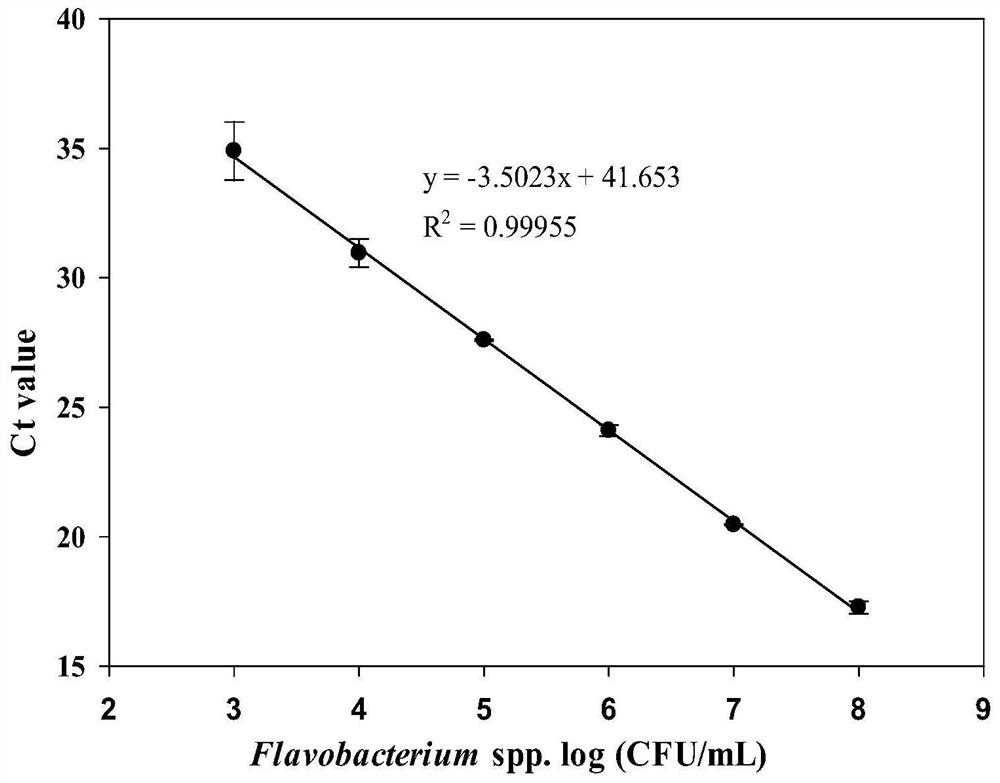
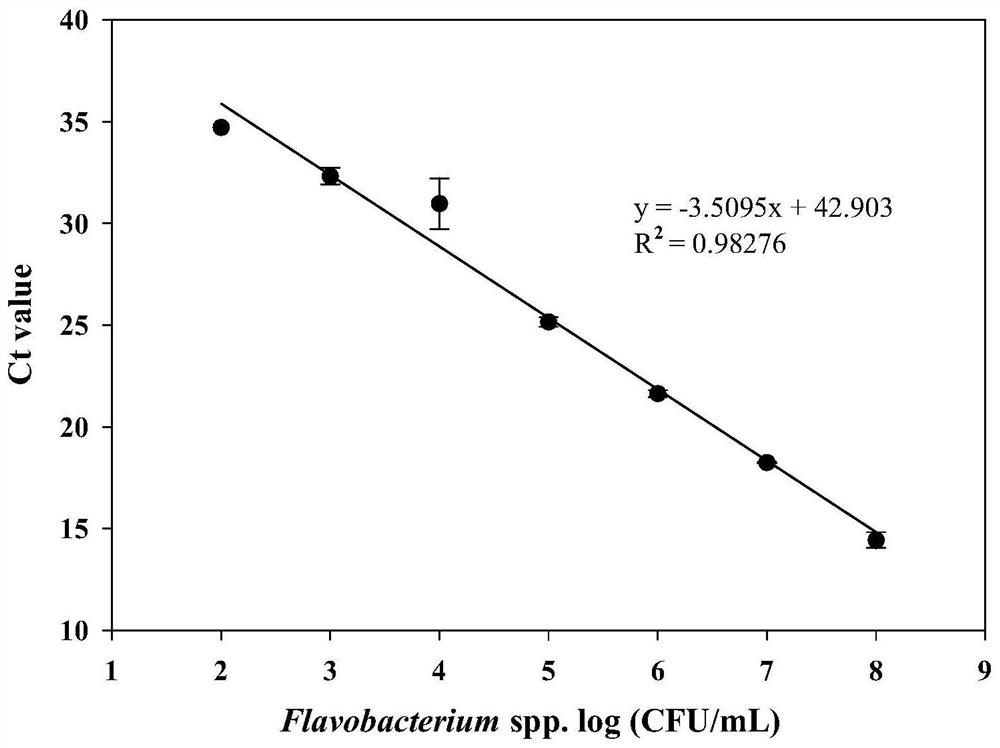
Molecular target for screening flavobacterium and quantitative detection method for flavobacterium
A technology for screening Flavobacterium and molecular targets, applied in the field of microbial inspection, can solve the problems of reporting and difficulties in molecular detection targets of Flavobacterium
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Screening to obtain the specific molecular target of Flavobacterium
[0030] According to the genome data of 271 strains of Flavobacterium and 40 strains of non-Flavobacteria in the NCBI website, pan-genome analysis was performed; Specific gene fragments of common Flavobacteria such as Flavobacterium sinensis, Flavobacterium meningitidis, Flavobacterium brevis, Flavobacterium halophilum, Flavobacterium psychrophilus), the nucleotide sequence of the gene fragment is as SEQ ID NO: 1.
[0031] The specific molecular targets of the Flavobacterium genus obtained by screening were all from Flavobacterium columnar 94-081, and the corresponding gene loci of the target gene fragment sequences are shown in Table 1 below.
[0032] Table 1 Genomic loci of specific molecular targets of Flavobacterium
[0033]
Embodiment 2
[0034] Embodiment 2 identifies the PCR method of Flavobacterium
[0035] 1) Primer design
[0036] Specific PCR amplification primers (including forward primers and reverse primers) were designed according to the sequence SEQ ID NO: 1 in Example 1, and the primer set sequence is shown in Table 2.
[0037] Table 2 Specific PCR detection primer set
[0038]
[0039] 2) the method for identifying Flavobacterium, the steps are as follows:
[0040] S1 DNA template preparation: the strains to be tested were enriched and cultured in LB liquid medium, and the bacterial genomic DNA was extracted using a commercial bacterial genomic DNA extraction kit as the template to be tested;
[0041] S2 PCR amplification: use the primer set 1 to perform PCR amplification of the sample DNA to be tested
[0042] ①PCR detection system:
[0043]
[0044] ②PCR amplification program:
[0045]
[0046] S3: Take the PCR amplification product for gel electrophoresis;
[0047] S4: Observe whe...
Embodiment 3
[0048] The specificity evaluation of embodiment 3 Flavobacterium PCR detection method
[0049] Take 5 strains of Flavobacterium and 44 strains of non-target Flavobacterium, and perform PCR detection according to the method in Example 2. Wherein, the S1 DNA template is prepared by extracting the genomic DNA of each bacterium; the S2 PCR amplification uses primer set 1. A blank control is set, and the template of the blank control is an aqueous solution without genome.
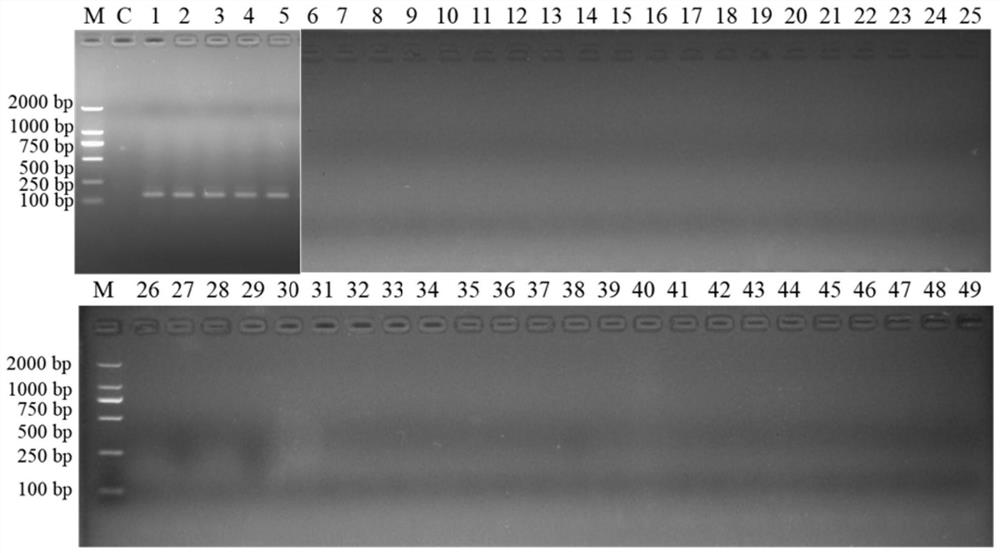
[0050] The strains and test results of the bacteria used are shown in Table 3 below. In the table, "+" in the test result column indicates positive, and "-" indicates negative. The results of electrophoresis of PCR products were as follows: figure 1 Shown; numbers 1 to 5 in the figure are 5 species of Flavobacteria; numbers 6 to 49 are non-target Flavobacteria; M is 2000Maker.
[0051] Table 3 Flavobacterium identification specificity evaluation test result of the present invention
[0052]
[0053]
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com