Application of integrated high-frequency gene loci of high and medium risk type HPV related to cervical cancer generation
A technology of integration sites and cervical cancer, applied in the field of molecular genetics, to achieve the effect of low false positive rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
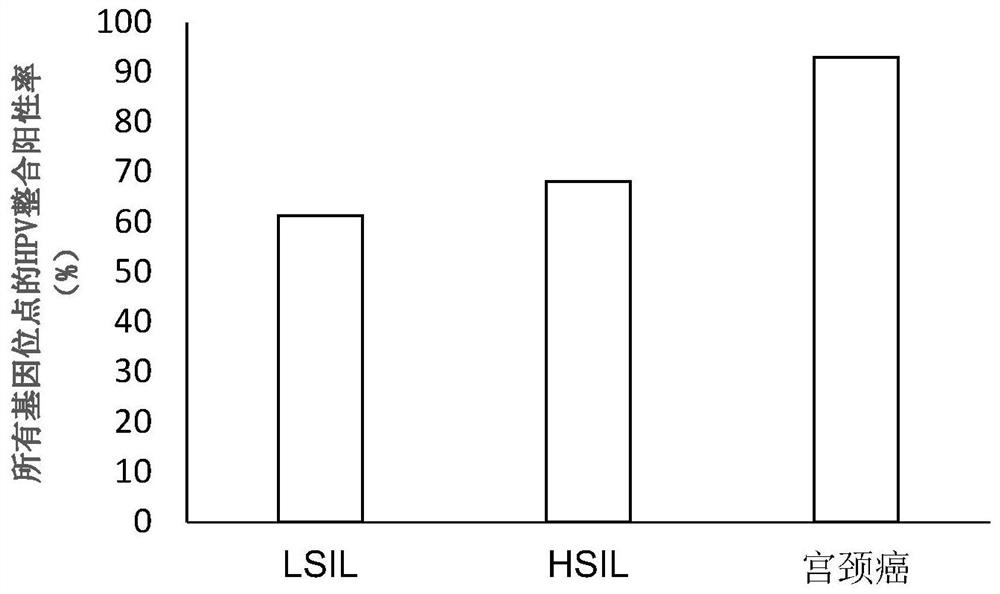
[0081] The cervical exfoliated cells of 988 LSIL patients, 358 HSIL patients, and 126 cervical cancer patients were selected as samples.
[0082] LSIL in the present invention refers to low-grade squamous intraepithelial lesion, and HSIL refers to high-grade squamous intraepithelial lesion, which are different pathological stages of cervical intraepithelial neoplasia.
[0083] 1. Centrifuge the exfoliated cells of the cervix for 5 minutes to collect the cells, discard the supernatant; add 200 μl PBS and 20 μl Proteinase K in sequence, shake and mix well;
[0084] 2. Add 200 μl of Buffer BCL, oscillate and mix well, bathe in 56°C water for 10 minutes, mix up and down several times during this period, and pass through the column for purification; add 150 μl of absolute ethanol, oscillate to mix, and briefly centrifuge to collect the liquid on the inner wall of the tube cap;
[0085]3. Put the FastPure gDNA Mini Columns II adsorption column in 2ml Collection Tubes, transfer the a...
Embodiment 2
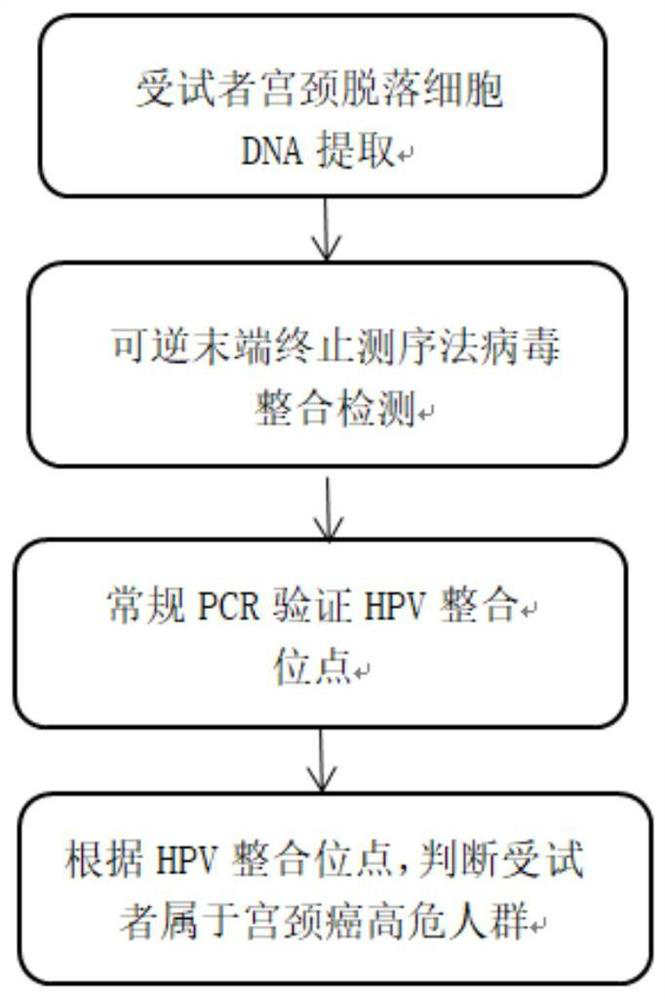
[0092] Detection of HPV integration sites in genomic DNA of cervical tissue by reversible end-termination sequencing.
[0093] 1. Virus integration detection of genomic DNA samples of cervical exfoliated cells: Design HPV full-length probes containing 18 subtypes (HPV16, HPV18, HPV26, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51, HPV52, HPV53, HPV56 , HPV58, HPV59, HPV66, HPV68, HPV73, HPV82), for the detailed content of the probe sequence, please refer to the applicant's patent application CN112195287A on November 12, 2020 - a human papillomavirus HPV typing and integration detection Probe sets and kits thereof;
[0094] Construct a DNA sequencing library according to the requirements of Illumina, use enzyme digestion to digest the sample genomic DNA into small fragments, and then repair and add A to the ends of these small fragments; ensure that the total amount of the constructed DNA library should be ≥ 1500ng, and The main peak of the library fragment length should be about 3...
Embodiment 3
[0099] The HPV integration sites were verified by PCR amplification and Sanger sequencing.
[0100] 1. PCR amplification
[0101] Design the upstream and downstream primers that can amplify the HPV integration site, and the primers are as shown in Table 2, and are prepared according to the system shown in Table 4 below:
[0102] Table 4 Primer System
[0103]
[0104] Mix each tube separately, put it into a standard PCR instrument for amplification, and perform amplification according to the procedures shown in Table 5 below:
[0105] Table 5 PCR amplification program
[0106]
[0107] 2. Send the amplified PCR product to Wuhan Qingke Biotechnology Co., Ltd. for Sanger sequencing.
[0108] (1) The PCR product obtained after amplification (content greater than 200ng, volume greater than 20ul) was sent to Wuhan Qingke Biotechnology Co., Ltd. for Sanger sequencing;
[0109] (2) Purify the PCR sample and add it to an ABI3730XL sequencer to detect the sequence of the PCR ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com