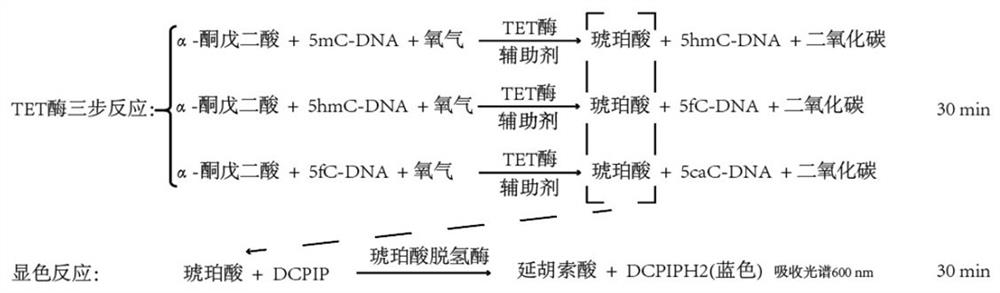
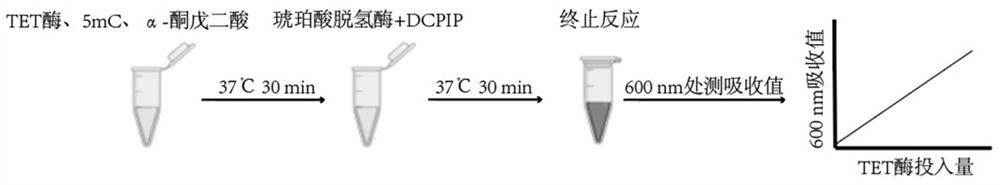
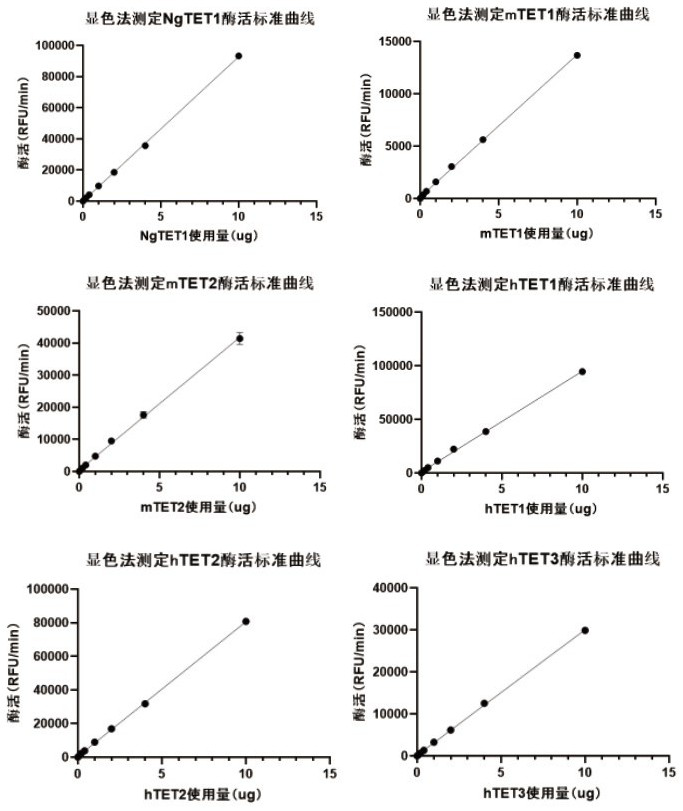
TET enzyme activity determination method and high-throughput screening method of TET enzyme activity micromolecule activator or inhibitor
A technology for enzyme activity determination and screening method, applied in the biological field, can solve problems such as time-consuming, high cost, complicated operation, etc., and achieve the effect of wide application range, high accuracy and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Determination of TET enzyme activity process and standard curve by chromogenic method.
specific Embodiment approach
[0041] In this example, we have published the measurement process of TET enzyme activity (such as figure 1 and figure 2 ), and the enzymatic activities of NgTET1(Active motif), mTET1(WiseGene), mTET2(Creative Biomart), hTET1(ActiveMotif), hTET2(Active Motif) and hTET3(Active Motif) were determined according to this process (such as image 3 ). The specific implementation is as follows:
[0042] Table 1
[0043] components Dosage 5mC DNA Standard 10ng TET enzyme reaction buffer 3 μL TET enzyme to be tested 0.1-10μg Add ddH2O to 30μL
[0044] React at 37°C for 30 min.
[0045]Table 2
[0046] components Dosage above reaction 30μL Succinate Dehydrogenase Reaction Buffer 4 μL SDHA 2 μg SDHB 2 μg Add ddH2O to 40μL
[0047] React at 37°C for 30 min. After the reaction, 5 uL of 35% trichloroacetic acid was added to terminate the reaction.
[0048] The peak absorption of the reactio...
Embodiment 2
[0050] Example 2: Comparison of three TET enzyme assay methods.
[0051] In this example, we compared the effect of three enzyme activity assay methods on TET enzyme activity: chromogenic method, LC-MS method and ELISA method.
[0052] Determination of TET enzyme activity by LC-MS method: phenol-chloroform extraction and ethanol precipitation were used to recover DNA, and LC-MS was used to measure the content ratio of 5mC, 5hmC, 5fC and 5caC in DNA (Hashimoto H , Pais J E , et al. Structure of aNaegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature, 2013, 506(7488):391-395.).
[0053] Determination of TET enzyme activity by ELISA: Use Epigentek's Epigenase 5mC-hydroxylase TET activity / inhibition assay kit (fluorescence method) to determine the enzyme specific activity of each TET protein according to the procedure in the manual.
[0054] The result is as Figure 4-Figure 9 As shown, compared with the LC-MS method and the ELISA method, the chromogenic met...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com