Primer probe combination and kit for plasmid quantification
A technology of primer probes and kits, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of low-copy template quantification, expensive consumables, cumbersome operations, etc., and achieve guaranteed Authenticity and traceability, avoiding false positive results, and high data reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Plasmid Quantification Kit Primer Probe Design
[0060] According to the AmpR and Kan gene sequences queried in the NCBI GeneBank database, according to the principles of TaqMan probe and primer design, the following two pairs of primer probes were optimized, and the sequences (5'-3') were as follows: wherein, the AmpR P probe was at the 5' The FAM fluorophore is labeled at the end, and the BHQ1 quencher is labeled at the 3' end; the Kan P probe is labeled with the VIC fluorophore at the 5' end, and the BHQ1 quencher is labeled at the 3' end.
[0061] AmpRF: CCAGTGCTGCAATGATACCG;
[0062] AmpRR: GGCTGGCTGGTTTATTGC;
[0063] AmpRP: FAM-ACCCACGCTCACCGGCTCCAGAT-BHQ1;
[0064] Kan F: AGCCAGTTTAGTCTGACCATC;
[0065] Kan R: GCCATATTCAACGGGAAACG;
[0066] Kan P: VIC-TCTGTAACATCATTGGCAACGCTACCT-BHQ2.
[0067] The above-mentioned primer probe sequence may also be a sequence with more than 85% homology with the above-mentioned sequence.
[0068] The above primers and probes...
Embodiment 2
[0070] Kit reaction system preparation
[0071] Prepare the reaction buffer for 100 tests according to the table below. The reaction buffer is divided into 20 μL for each reaction, 5 μL of the template is added, and the total volume is 25 μL.
[0072]
[0073]
Embodiment 3
[0075] Preparation of standard and reference substances
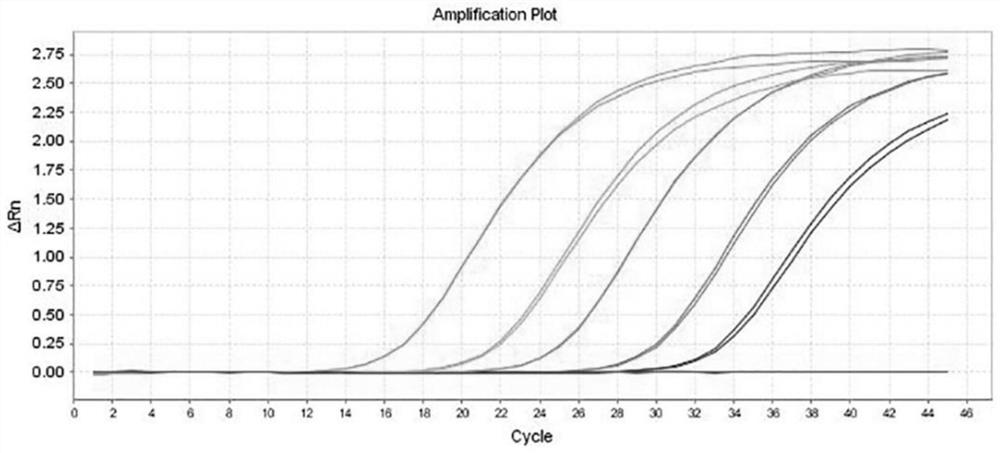
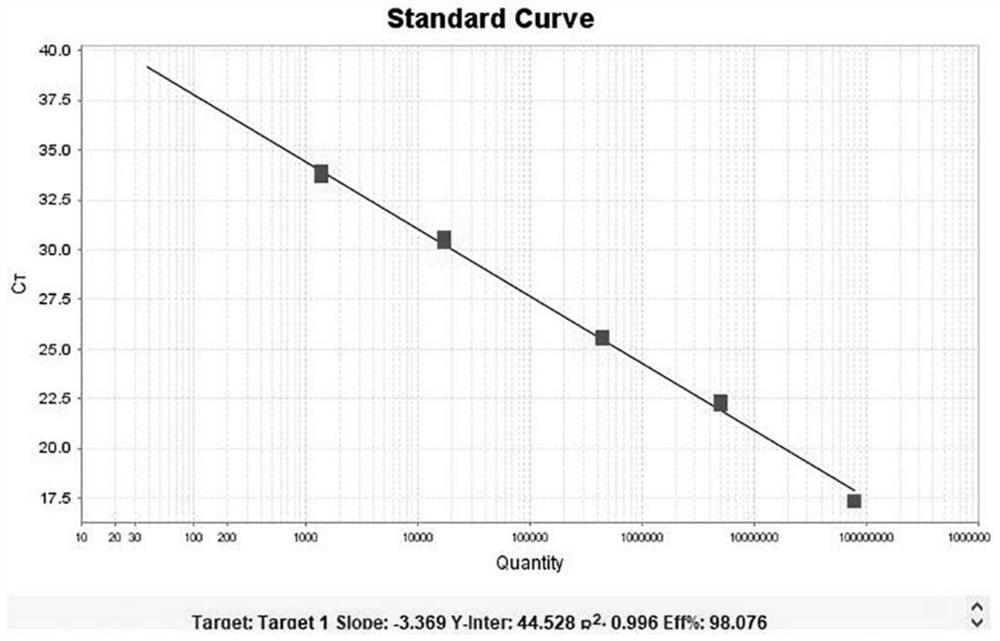
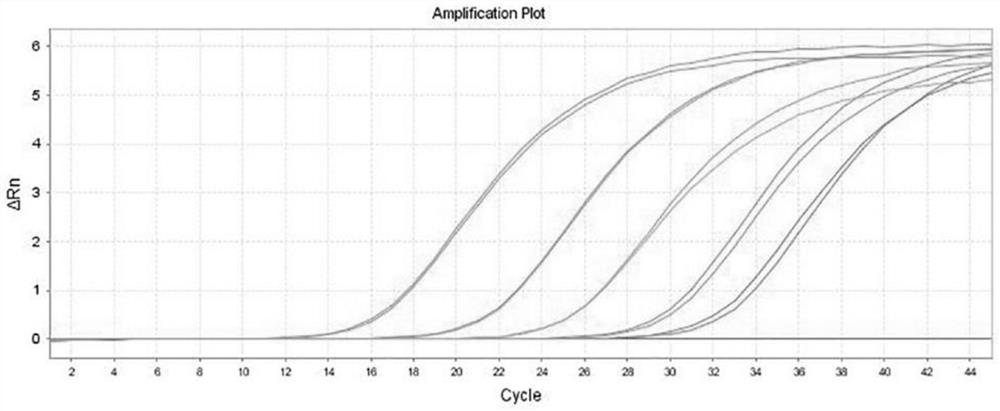
[0076] 在pUC57-AmpR质粒载体上插入含有卡那霉素抗性基因的序列,插入序列为:AAATTCCGTCAGCCAGTTTAGTCTGACCATCTCATCTGTAACATCATTGGCAACGCTACCTTTGCCATGTTTCAGAAACAACTCTGGCGCATCGGGCTTCCCATACAAGCGATAGATTGTCGCACCTGATTGCCCGACATTATCGCGAGCCCATTTATACCCATATAAATCAGCATCCATGTTGGAATTTAATCGCGGCCTCGACGTTTCCCGTTGAATATGGCTCAT,酶切位点为KpnI / SalI。 After constructing the vector, it was transferred into Escherichia coli for expanded culture, and the plasmid was extracted for digital PCR quantification. A total of 5 standard products were quantified, and the concentrations were: A 8*10 7 copies / mL, B 5*10 6 copies / mL, C 4.5*10 5 copies / mL, D 1.7*10 4 copies / mL, E 1.4*10 3 copies / mL.
[0077] Take the quantitative concentration as 5*10 6 The copies / mL plasmid is used as the positive control of the kit, and TE Buffer is used as the negative control of the kit.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com