Whole-chromosome genotyping chip for synchronously detecting multiple birth defect genetic diseases and method and application of whole-chromosome genotyping chip
A technology for chromosomal genes and birth defects, applied in biochemical equipment and methods, genomics, sequence analysis, etc., to achieve high operability, improved diagnosis, and high timeliness.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
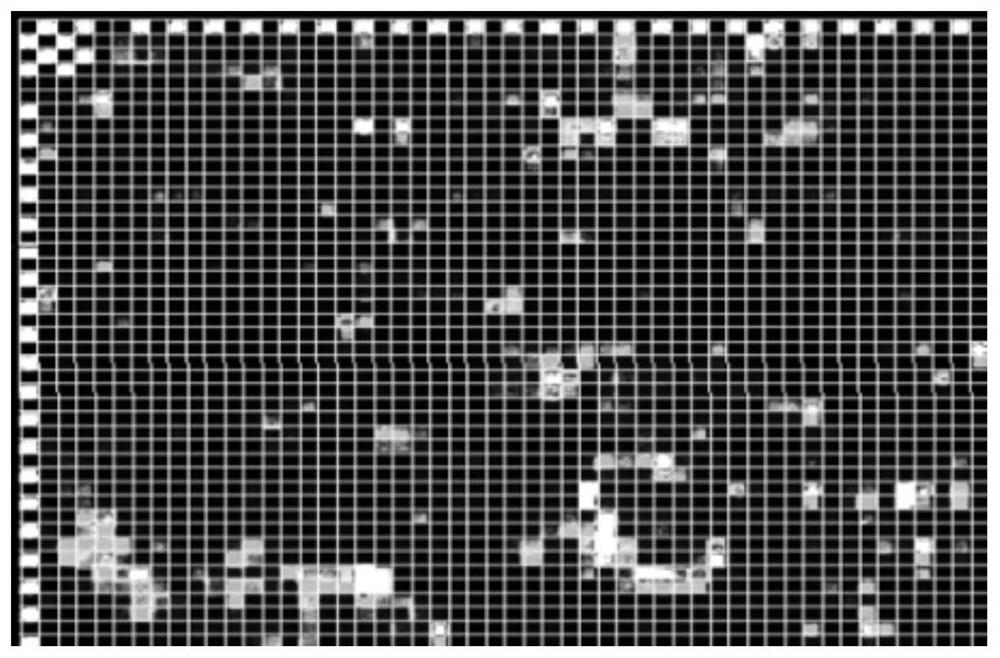
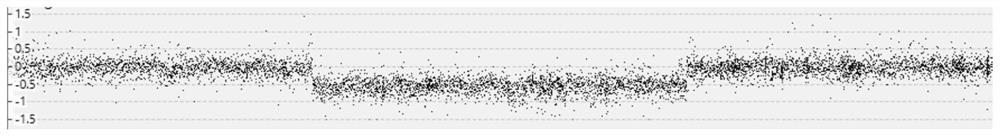
Image
Examples
Embodiment 1
[0102] A whole-chromosomal genotyping chip for synchronous detection of multiple birth defects and genetic diseases in this embodiment, the chip includes a genome_framework probe set and a special_gene probe set.
[0103] The genome_framework probe set includes skeleton probes designed for autosomes and targeted chromosomes; skeleton probes reduce / remove the head and end regions of each chromosome, centromere regions and other gene-free region probes;
[0104] The genome_framework probe set includes probes designed for 16 gene copy number variation (CNV) regions at the single gene level. The 16 single genes include VHL, QDPR, SMN1, CYP21A2, HBB, PTS, PAH, ATP7B, GCH1, HBA1, HBA2, NF1, PHEX, DMD, OTC, MECP2;
[0105] The genome_framework probe set includes probes designed for 350 pathogenic haploinsufficiency (HI) genes and gene copy number variation (CNV) regions of triple dose-sensitive (TS) genes, pathogenic haploinsufficiency ( HI) genes and triple dose-sensitive (TS) gene...
Embodiment 2
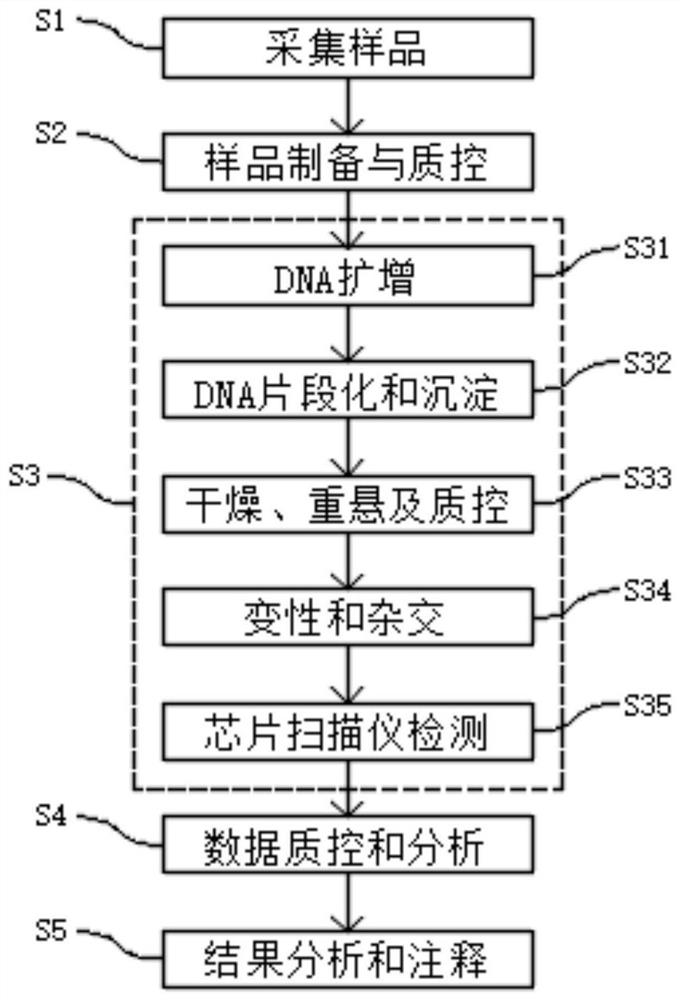
[0205] A method for synchronously detecting multiple birth defect genetic diseases CNV and SNV in the example of this embodiment includes the following steps, such as figure 1 as shown,
[0206] S1: Collect samples: a total of 96 samples, 54 males and 42 females, 48 of which are verified patients carrying CNV, and the other 48 are verified positive verification samples of the invention .
[0207] S2: Sample preparation and quality control. After the DNA extraction of the verification sample, the quality control is carried out through gel electrophoresis, Nanodrop, etc. to ensure that each DNA sample has no degradation, no impurity contamination, and high purity. The optical density (Optical Density, OD) 260 / 280nm ratio is between 1.8 and 2.0, and the OD 260 / 230nm ratio is between 1.5 and 2.0. Samples that do not meet any of the conditions need to be purified and other treatments; the gel loading dye is Invitrogen TM TrackIt Cyan / Orange Loading Buffer (Invitrogen P / N 10482-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com