MNP marker site of herpes simplex virus, primer composition, kit and application of MNP marker site
A herpes simplex virus and primer combination technology, applied in the biological field, can solve the problems of high typing error rate, low accuracy, unacceptable sequencing cost, etc., and achieve high-sensitivity detection and discrimination, high accuracy and detection and discrimination. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
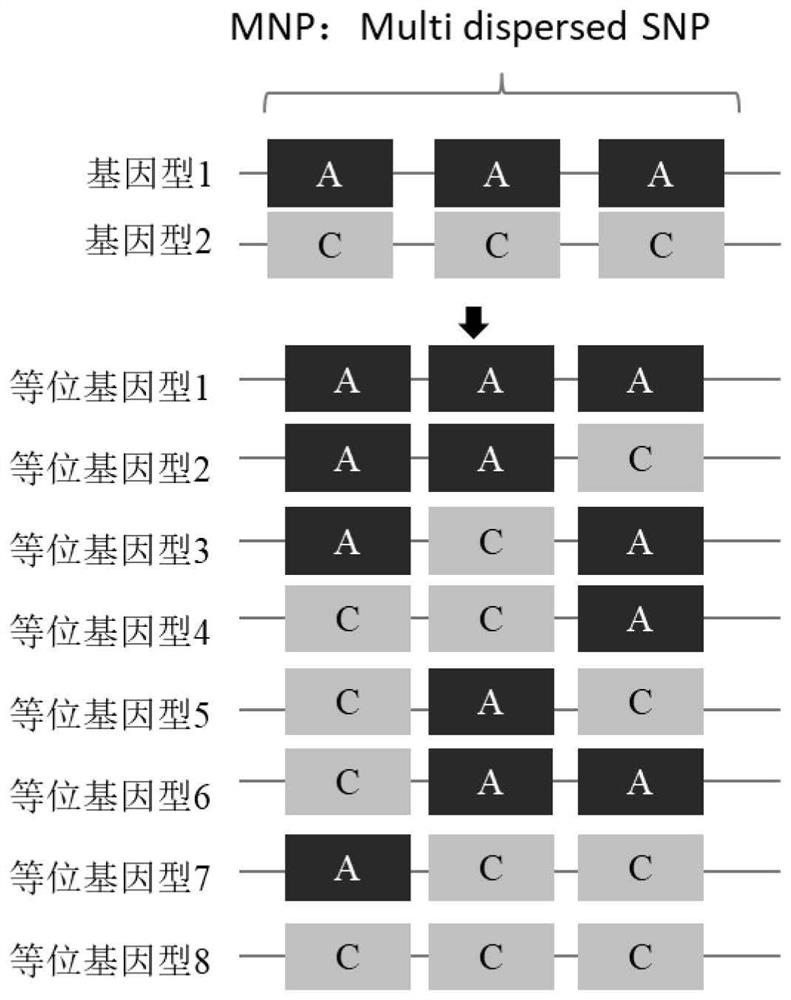
Image
Examples
Embodiment 1
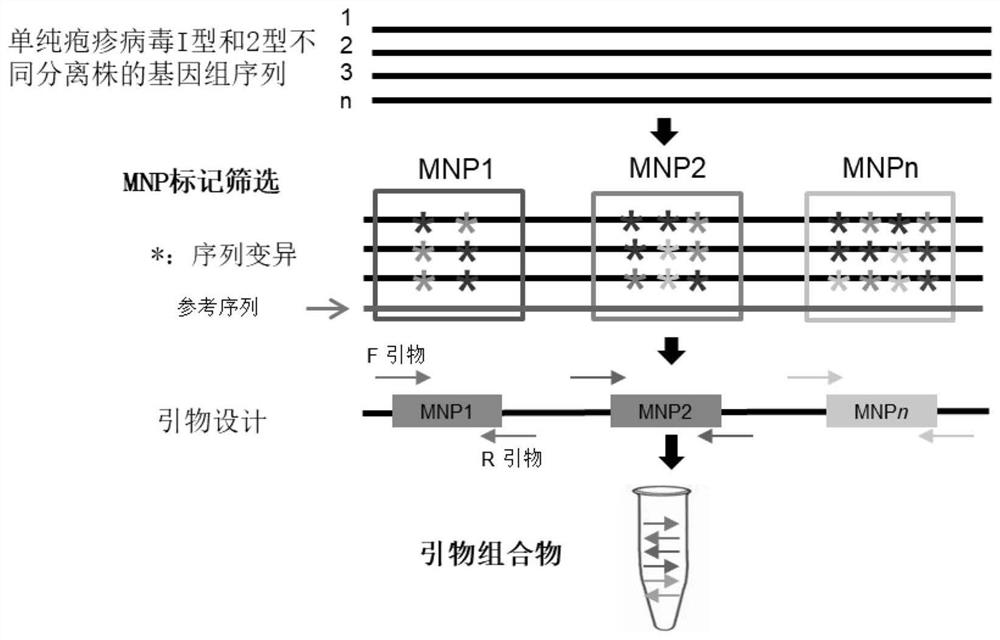
[0042] Example 1. Screening of herpes simplex virus MNP marker sites and design of multiple PCR amplification primers
[0043] S1. Screening of herpes simplex virus MNP marker sites
[0044] Based on the genome sequences of 17,960 HSV isolates with different serotypes published on the Internet, 7 HSV-1-specific and 7 HSV-2-specific, total 14 MNP marker sites were obtained through sequence alignment. For species that do not have genomic data online, the genome sequence information of representative isolates of the microbial species to be detected can also be obtained by high-throughput sequencing, where high-throughput sequencing can be whole genome or simplified genome sequencing. In order to ensure the polymorphism of the selected marker, the genome sequences of at least 10 genetically representative isolates are generally used as a reference. The 14 MNP marker sites screened are shown in Table 1:
[0045] Table 1 - The MNP marker site and the starting position of the detec...
Embodiment 2
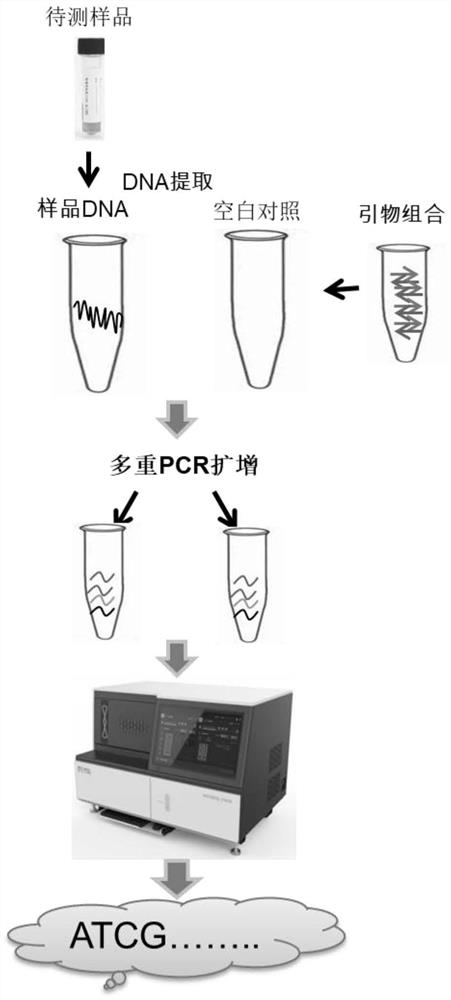
[0059] Embodiment 2, MNP markers and primers identify the performance evaluation and threshold setting of herpes simplex virus
[0060] In this example, the nucleic acids of herpes simplex virus type 1 and type 2 with known copy numbers were added to the genomic DNA, respectively, to prepare 3 kinds of herpes simplex virus simulations of 1 copy / reaction, 10 copies / reaction and 100 copies / reaction sample. At the same time, an equal volume of sterile water was set as a blank control. 4 samples were tested for each virus, and 3 replicate libraries were constructed for each sample every day for 4 consecutive days, that is, 12 sets of sequencing data were obtained for each sample of each virus. Table 3 shows the data analysis results of herpes simplex virus type 1 . The reproducibility, accuracy and sensitivity of the detection method were evaluated according to the number of MNP-labeled sequencing fragments and sites of herpes simplex virus type 1 and type 2 detected in blank co...
Embodiment 3
[0088] Embodiment 3. Detection of genetic variation among herpes simplex virus strains
[0089] 3 HSV-1 and 2 strains provided by the Hubei Provincial Center for Disease Control and Prevention were tested by the kit. The samples were named S1-S6 in turn. The average coverage of each marker for each sample was sequenced. up to 1202 times. Among them, the three strains of HSV-1 are three progeny strains of the same strain preserved in different periods. The results are shown in Table 6, and 7 MNP markers could be detected for each strain (Table 6). The fingerprints of the 6 strains were compared pairwise, and the 6 strains could be divided into 2 subtypes, S1-S3 belonged to one subtype, and S4-S6 belonged to one subtype. S1-S3 are three progeny strains of the same strain preserved in different periods. S1, S2, and S3 all have major genotype differences in two markers (Table 6), indicating that there is inter-strain variation.
[0090] Table 6-6 Detection and Analysis of Herpe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com