Method for constructing Eriocheir sinensis protein interaction network
A Chinese mitten crab and protein interaction technology is applied in the field of building protein interaction networks, which can solve the problems of huge differences in protein interaction networks, and achieve the effects of simple and easy operation, high quality and wide sources.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
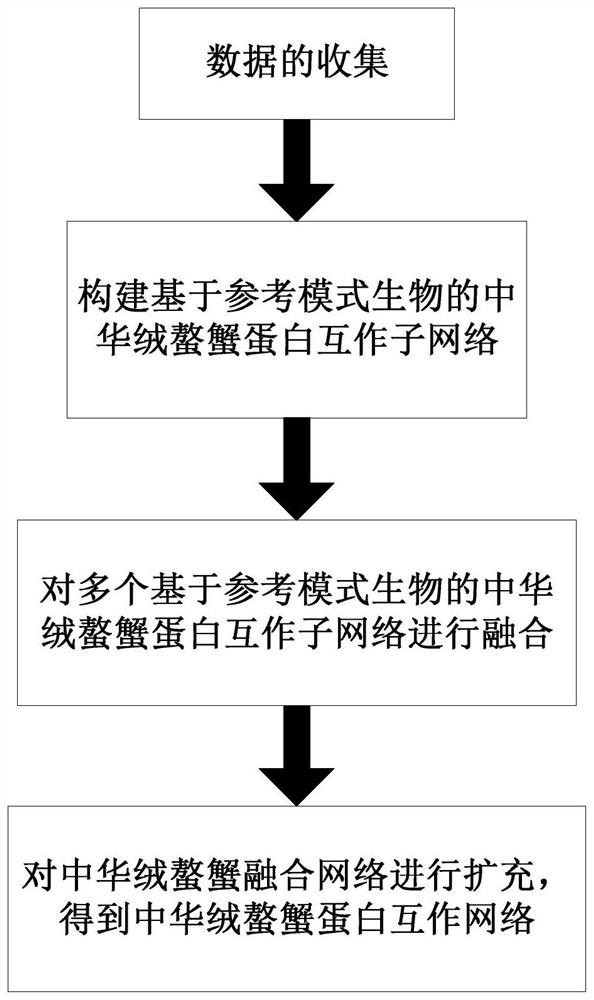
[0075] Taking the transcriptome of Chinese mitten crab as the source of gene sequences, and selecting Drosophila, nematode, human, house mouse, brown mouse, and yeast as reference model organisms, a protein interaction network of Chinese mitten crab was constructed. The network contains 8225 proteins and 148524 protein interactions. The specific method is as follows:
[0076] Step 1: Data Collection:
[0077](1) The gene sequence data of Eriocheir sinensis is taken from the transcriptome data of the GEO database. The data includes the transcriptome data of five tissues of Eriocheir sinensis gill, eye stalk, thoracic nerve mass, hepatopancreas, and muscle. Contains 246,232 gene sequences and their functional annotations;
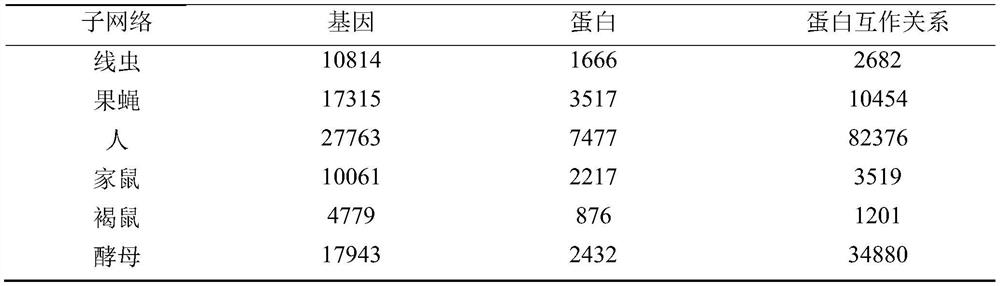
[0078] (2) Obtaining 22,006 Drosophila protein sequences, 26,618 nematode protein sequences, 70,236 human protein sequences, 50,694 house mouse protein sequences, 29,982 brown mouse protein sequences, and 6,721 yeast protein sequences from the Uniprot datab...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com