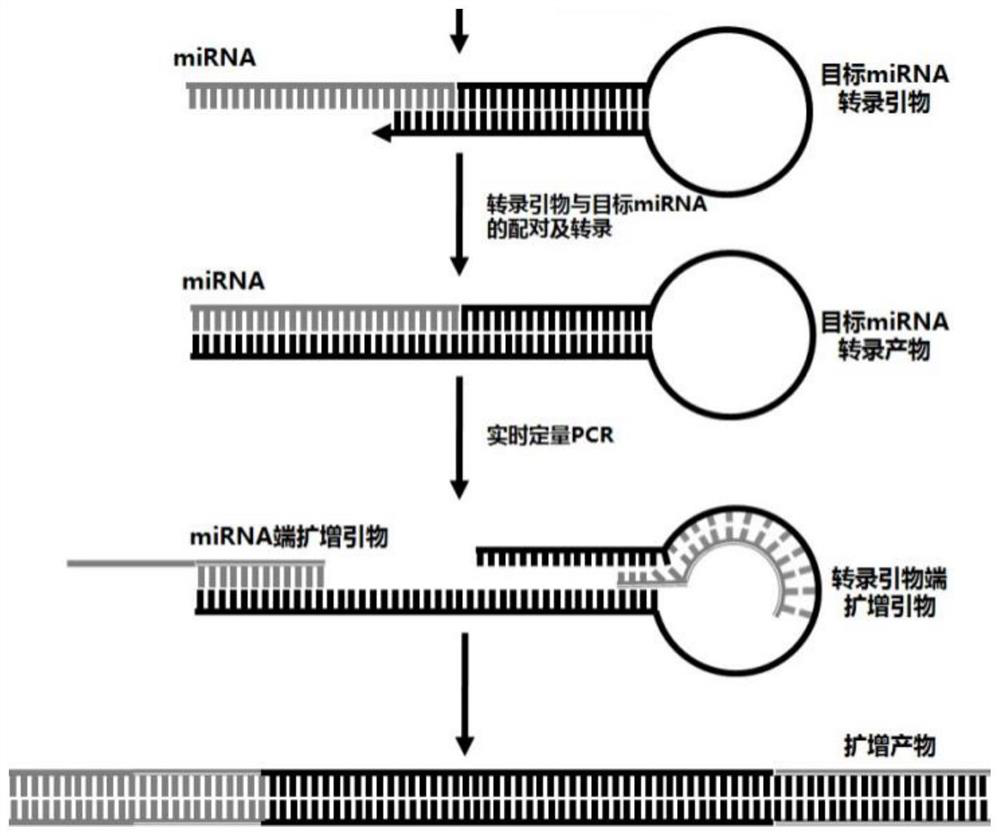
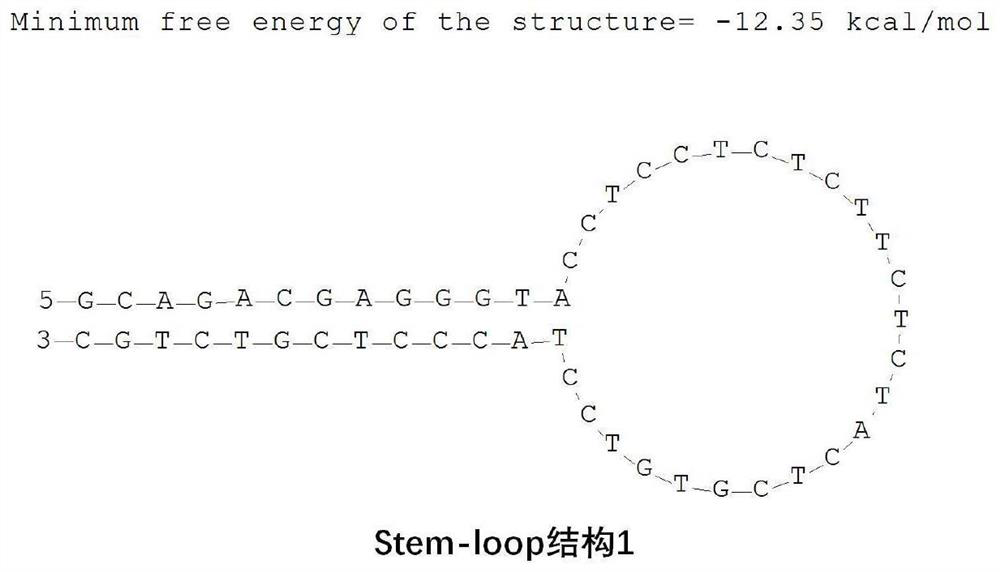
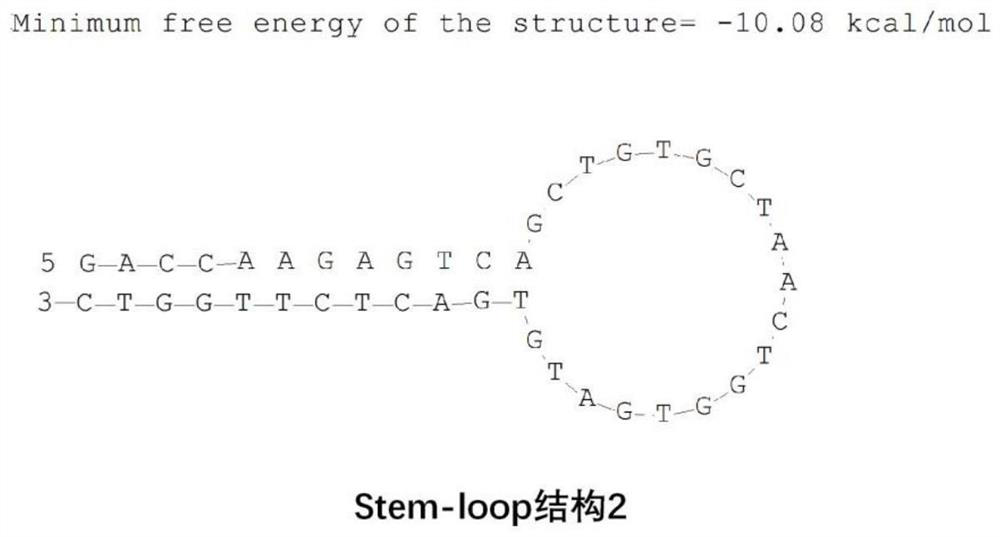
Skin repair related miRNA marker and application thereof
A skin repair and marker technology, applied in the field of kits and miRNA detection reagents, to achieve the effect of great practical significance and practical value, high accuracy and high degree of automation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Example 1: Skin injury modeling and skin collection
[0080] Nine adult female mice with similar body weights were selected and divided into 3 groups. Using a hole puncher to create wounds on the back skin of the mice respectively, the skin was collected as a 0-day sample. Then, the wound edge skins of the three groups of mice were collected on the 7th day, 14th day and 21st day of wound modeling respectively. After the skin was collected, it was quick-frozen in liquid nitrogen and transferred to -80°C for storage until RNA extraction.
Embodiment 2
[0081] Example 2: Extraction of total RNA from skin
[0082] Take 50-100 mg of skin samples, add 6 metal balls with a diameter of 2 mm and 2 metal balls with a diameter of 5 mm and 1 mL of Trizol reagent from Invitrogen Company, fasten the cap of the centrifuge tube, and use the Jingxin tissue grinder to grind the sample at 70 Hz and 90 s. Cryogenic grinding; after placing the ground sample tubes in a refrigerator at 4°C for 5 min, add 200 μL of chloroform to each tube, fasten the cap of the centrifuge tube, shake by hand, place in a refrigerator at 4°C for 5 min, and centrifuge at 12000 rpm for 15 min at 4°C The liquid in the tube is divided into 3 clear layers; prepare a new 1.5 mL centrifuge tube without nuclease and number it, add 500 μL of isopropanol to each tube, take out the centrifuged specimen, and carefully pipette 500 μL of the upper transparent liquid to the corresponding numbered tube. In the centrifuge tube, fasten the cap of the centrifuge tube, vortex for 5s, ...
Embodiment 3
[0083] Example 3: High-throughput sequencing of miRNAs
[0084] The miRNA library was constructed and sequenced, followed by the identification of differentially expressed genes, and the P value was adjusted using the Benjamini & Hochberg method. By default, a corrected P-value of 0.05 or 0.01 was set as the threshold for significant differential expression. On the target gene candidates of differentially expressed miRNAs, Gene Ontology (GO) enrichment analysis was used. GOseq-based Wallenius non-central hypergeometric distribution can adjust gene length bias for GO enrichment analysis. Potential functions of predicted miRNAs by KEEG pathway enrichment. Statistical enrichment of target gene candidates in the KEGG pathway was tested using KOBAS software. The candidate miRNAs related to the wound repair process were determined, and the final screening found that miRNAs miR-7a-5p, miR-335-5p and miR-501-3p played a key role in the wound repair process.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com