Molecular identification method for sponge associated dominant bacterium constitution and host specific bacterium
A sponge symbiosis, host-specific technology, applied to the identification of the gene level, the composition of sponge symbiosis dominant bacteria and the molecular identification of host-specific bacteria, can solve the problem of not revealing the symbiotic microbial composition and structure of different sponges. Diversity and host-specific bacteria, time-consuming, complex operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0016] The technical solutions of the present invention will be further described below in conjunction with the accompanying drawings and embodiments.
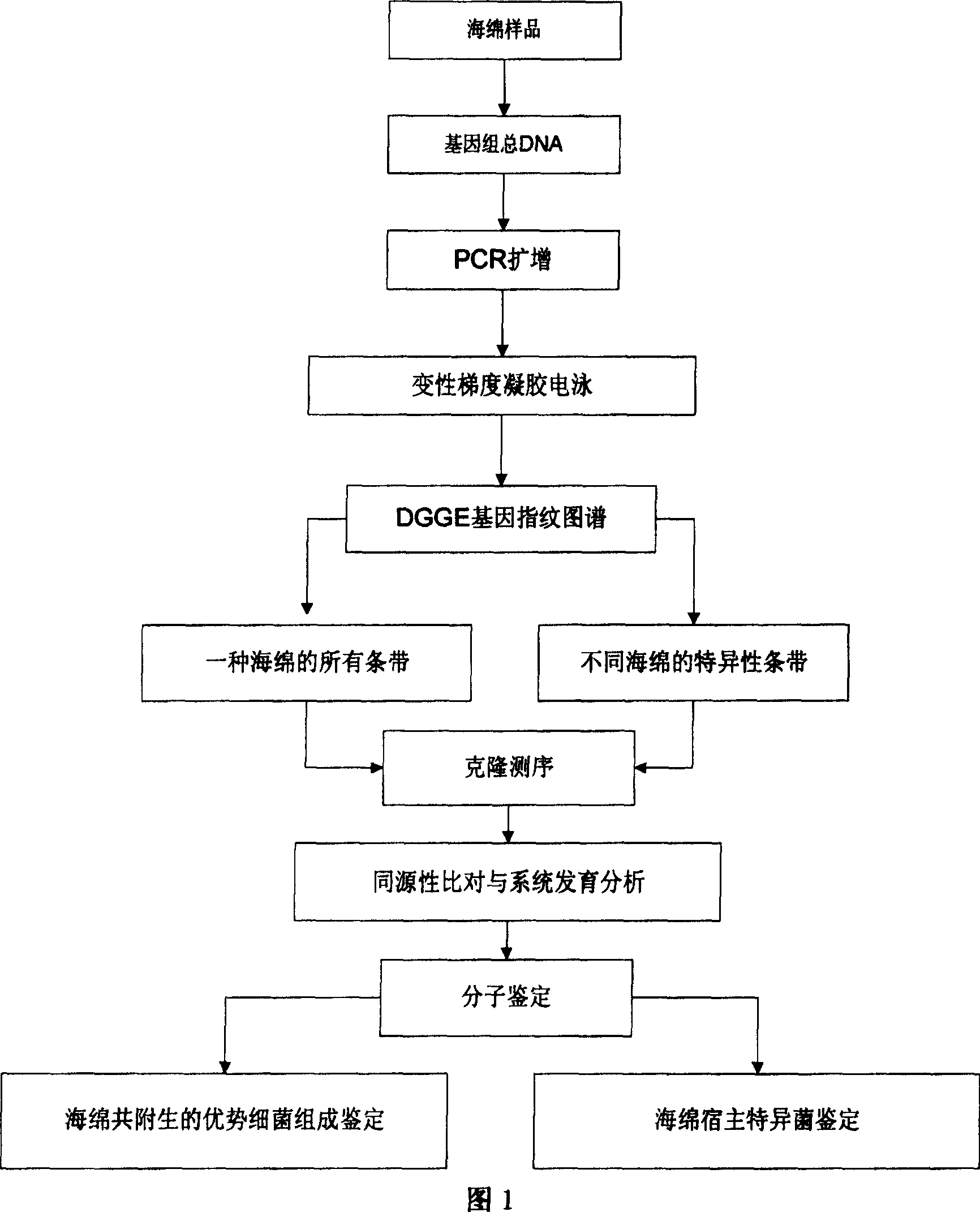
[0017] The flow process of the method of the present invention is shown in Figure 1, at first obtain sponge sample, extract genome total DNA, utilize polymerase chain reaction to amplify and obtain the 16S rDNA fragment of bacterium, carry out denaturing gradient gel electrophoresis to construct fingerprint atlas, for a kind of sponge Select all the bands, and find out the specific bands of a sponge by comparing the differences in the fingerprints constructed by denaturing gradient gel electrophoresis of different sponges. These bands are tapped, recovered, cloned and sequenced, and compared with the gene bank for homology And phylogenetic tree analysis to complete the molecular identification of bacteria, thus revealing the dominant bacterial composition of sponge symbiosis and the molecular identification of sponge host-speci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com