Tobacco strong nuclear matrix binding sequence, separating identification method and use thereof
A nuclear matrix combination and sequence technology, applied in the field of tobacco strong MAR sequence and its isolation and identification, to achieve high expression activity, enhanced high-efficiency expression, and improved yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Cloning of a tobacco strong MAR sequence
[0043] a. PCR Reagents and Conditions for Polymerase Chain Reaction (PCR)
[0044] The template DNA of the PCR reaction is tobacco genome DNA. Tobacco (Nicotiana tabacum L.cv NC89) was grown in the greenhouse, and its leaves were used for the extraction of genomic DNA, which was extracted by the CTAB method
[0045] First mix the following reagents together:
[0046] 10×Taq DNA polymerase buffer 5 μl
[0047] Template DNA (100 ng / μl) 1 μl
[0048] Forward primer (100 nmol / L) 1 microliter
[0049] Reverse primer (100 nmol / L) 1 microliter
[0050] Deoxynucleotide mixture (dNTPs, 100 nmol / L) 1 μl
[0051] Taq DNA polymerase (5 units / μl) 0.5 μl
[0052] Sterilized redistilled water 30.5 microliters
[0053] Total volume 50 µl
[0054] The conditions of the PCR reaction were as follows: denaturation at 94°C for 5 minutes, followed by the following cycles: 94°C for 40 seconds, 46°C for 50 seconds, 72°C for 90 seco...
Embodiment 2
[0066] Example 2: In vitro binding experiment of MAR sequence and nuclear matrix
[0067] a. Suspension culture Rice seeds were used to induce callus on Ms modified medium (yeast powder 0.3%, 2,4-D 2 mg / L, 6 BA 0.5 mg / L). The loose and friable callus was picked and subcultured in AA (2,4-D 1 mg / L) liquid medium at 26° C., 110 rpm, every 9 days.
[0068] b. Extraction of cell nuclei Get 10 ml of suspension cells (cell volume) cultured to the 7th day, treat in 20 ml of enzymolysis solution containing 2% cellulase RS, 1% isolated enzyme and 0.2% pectinase Y 23, use Homogenize with a full glass homogenizer until it is ground to a single cell, pass through the 22μm and 15μm stainless steel mesh to sieve the coarse suspension of nuclei, centrifuge at 5000g for 20min, collect the precipitate, and use 60% and 30% Percoll 1000g discontinuous density Gradient centrifugation to collect the precipitate. 15OD(A per ml 260 ) was suspended in 24ml preservation solution, frozen in liquid n...
Embodiment 3
[0071] Embodiment 3: Construction of plant expression vector
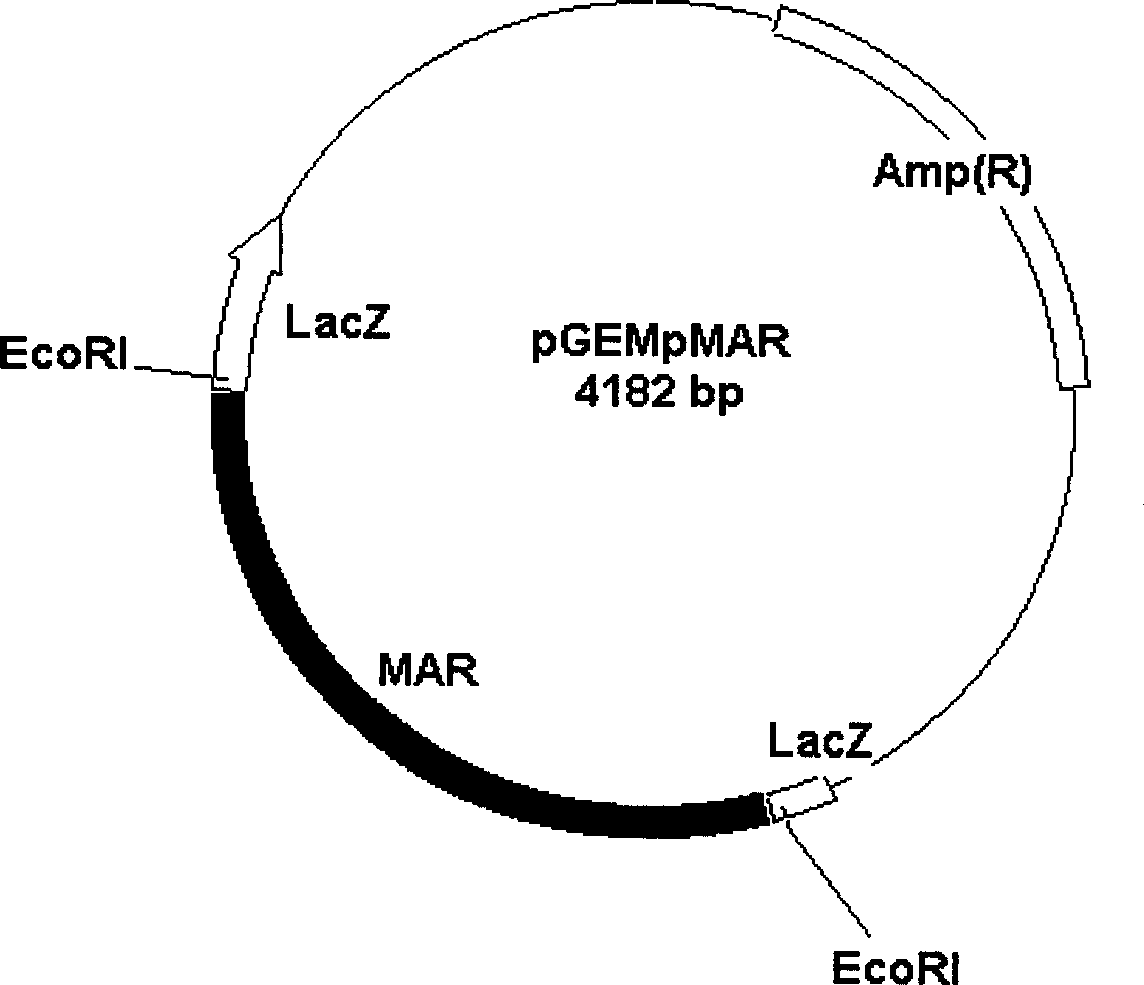
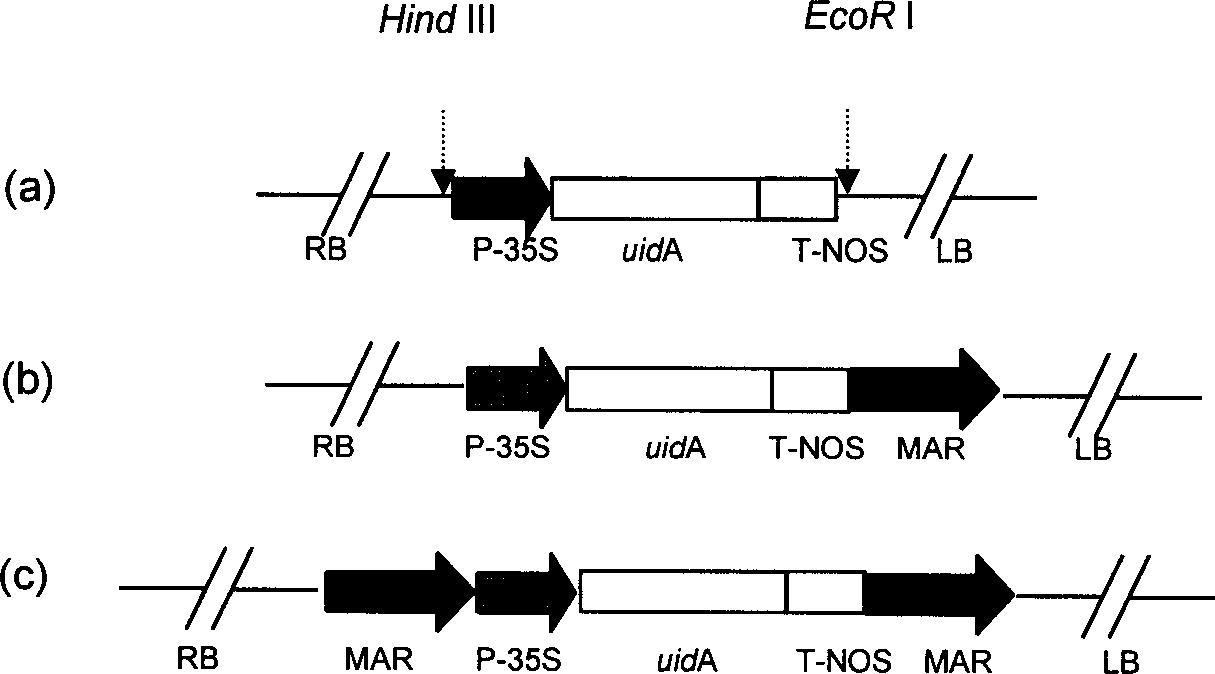
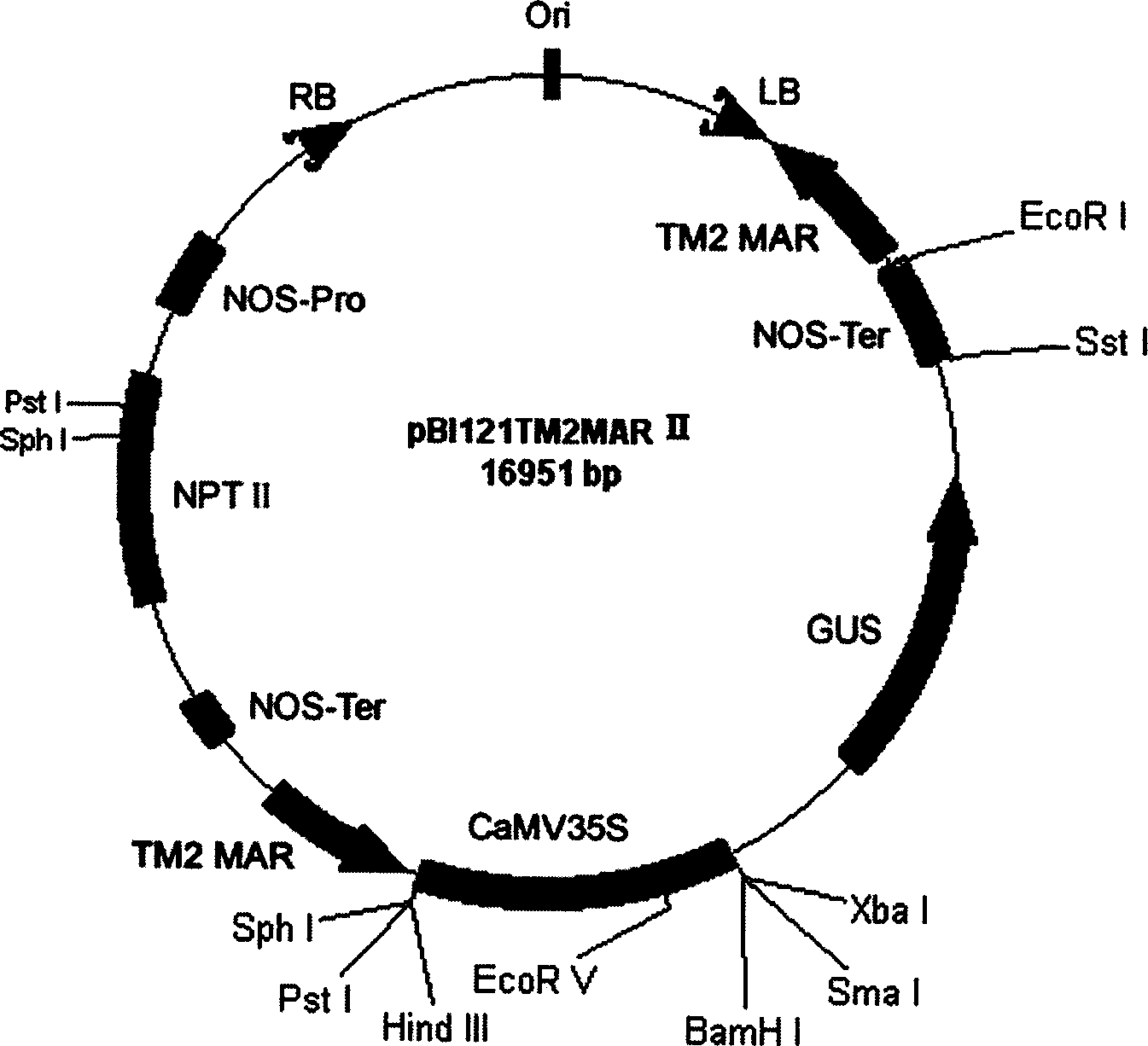
[0072] The MAR sequence was inserted into pBI121 ( figure 2(a)) Both sides of the gus gene expression cassette. MAR was excised from the plasmid pGEMpMAR with EcoR I, and a part was inserted into the EcoR I restriction site on the pBI121 treated with alkaline phosphatase to obtain the plasmid pBI121MAR I ( figure 2 (b)). The direction of insertion of the fragment was identified by PCR amplification using the gus upstream primer (ATGTTACGTCCTGTAGAAACCCCA) and the 3' end-specific primer of MAR. The expression vector pBI121MAR I inserted into the MAR was screened out, digested with Hind III and then dephosphorylated with alkaline phosphatase, and then the other part of the MAR with the sticky end of EcoR I was filled with Klenow enzyme and connected with the Hind III linker. Inserted into the HindIII restriction site of pBI121MAR I after Hind III digestion to generate pBI121MARII ( figure 2 (c)), using the 35S ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com