Process of preparing D-amino acid oxydase
An oxidase and amino acid technology, applied in the field of bioengineering, can solve problems such as difficult separation and achieve high-efficiency expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1 Obtaining mutant D-amino acid oxidase gene
[0018] According to the known DAAO gene sequence of Trigonopsis Vriablis and the codons corresponding to histidine, the mutation primers were designed as follows:
[0019] 5'primer 5' GGATCC ATGCACCATCATCATCATCAT ATGGCTAAAA TCGTTGTT
[0020] 3'primer 5' GCGGCCGC The TTAATGATGATGATGATGATG AAGGTTT GGACGAGTAAG template gene is derived from the plasmid pPIC3.5K-DAAO described in the patent CN 1385521A, and the DAAO gene is amplified by high-fidelity PCR (Pfu enzyme, purchased from the company), and the 5' and 3' ends of the coding sequence are respectively A Histag sequence that continuously encodes 6 histidines was introduced, and enzyme cleavage sites corresponding to NotI and BamHI were respectively introduced. The PCR reaction conditions are: 94°C for 5min, 1 cycle; 94°C for 30s, 40°C for 30s, 72°C for 75s, 5 cycles; 94°C for 30s, 58°C for 30s, 72°C for 75s, 30 cycles; finally, 72°C for 10min , 1 cycle.
[...
Embodiment 2
[0022] Example 2 Blunt-end cloning of mutant DAAO gene into plasmid pBluescript(SK+)
[0023] Plasmid pBluescript (SK+) was linearized with SmaI and used as a vector for blunt-end cloning. The mutant DAAO gene fragments recovered from the gel were ligated in vitro with the vector, and the reaction system (20 μl) was as follows:
[0024] pBluescript (SK+) 2μl
[0025] PCR product 10μl
[0026] 10x ligation buffer 2 μl
[0027] T4 DNA Ligase (1U / μl) 1μl
[0028] Water 5μl
[0029] The mixture was reacted overnight at 16°C. The obtained ligation product was transformed into E.coli TG1 competent cells, spread on LB plates containing ampicillin resistance, and the white transformants were selected to extract plasmid DNA, and the obtained DNA was proved to be pSK-DAAO by PCR and enzyme digestion.
Embodiment 3
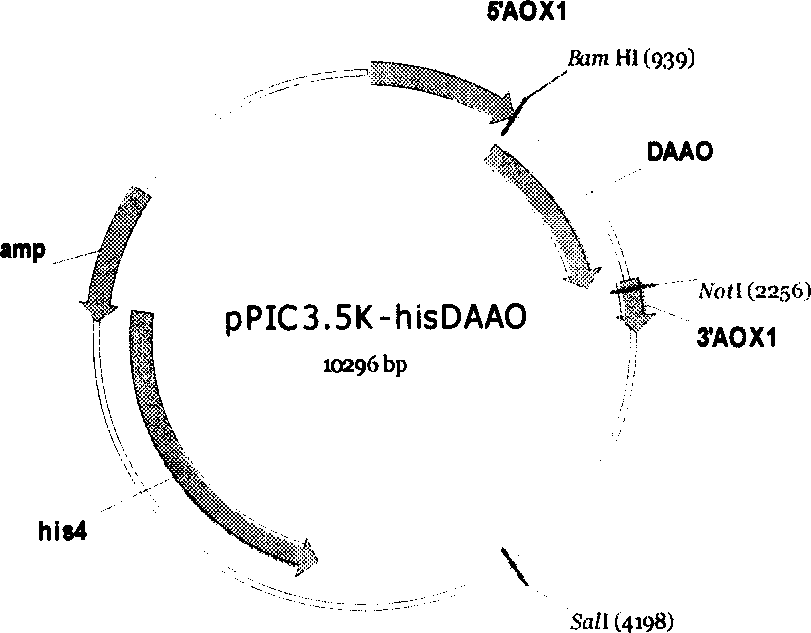
[0030] Example 3 Construction of expression plasmid pPIC3.5K-hisDAAO
[0031] Digest pSK-DAAO with NotI and BamHI, perform electrophoresis on 1% agarose gel, recover a fragment of about 1.3Kb from the gel, and then ligate it with the pPIC3.5K plasmid that has undergone the same double digestion in vitro. The reaction system is as follows:
[0032] pPIC3.5k (NotI and BamHI cut) 2μl
[0033] DAAO gene fragment 15μl
[0034] 10x ligation buffer 2μl
[0035] T4 DNA ligase 1μ
[0036] The above mixture was reacted overnight at 16°C. The resulting ligation product was transformed into E.coli TG1 competent cells, spread on LB plates containing ampicillin resistance, and the transformants were selected to extract plasmid DNA. The obtained DNA was proved to be pPIC3.5K-DAAO by PCR and enzyme digestion.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com